Masahiro Takatsuka
Graph Construction using Principal Axis Trees for Simple Graph Convolution
Mar 01, 2023Abstract:Graph Neural Networks (GNNs) are increasingly becoming the favorite method for graph learning. They exploit the semi-supervised nature of deep learning, and they bypass computational bottlenecks associated with traditional graph learning methods. In addition to the feature matrix $X$, GNNs need an adjacency matrix $A$ to perform feature propagation. In many cases the adjacency matrix $A$ is missing. We introduce a graph construction scheme that construct the adjacency matrix $A$ using unsupervised and supervised information. Unsupervised information characterize the neighborhood around points. We used Principal Axis trees (PA-trees) as a source of unsupervised information, where we create edges between points falling onto the same leaf node. For supervised information, we used the concept of penalty and intrinsic graphs. A penalty graph connects points with different class labels, whereas intrinsic graph connects points with the same class label. We used the penalty and intrinsic graphs to remove or add edges to the graph constructed via PA-tree. This graph construction scheme was tested on two well-known GNNs: 1) Graph Convolutional Network (GCN) and 2) Simple Graph Convolution (SGC). The experiments show that it is better to use SGC because it is faster and delivers better or the same results as GCN. We also test the effect of oversmoothing on both GCN and SGC. We found out that the level of smoothing has to be selected carefully for SGC to avoid oversmoothing.
Random projection tree similarity metric for SpectralNet
Feb 25, 2023Abstract:SpectralNet is a graph clustering method that uses neural network to find an embedding that separates the data. So far it was only used with $k$-nn graphs, which are usually constructed using a distance metric (e.g., Euclidean distance). $k$-nn graphs restrict the points to have a fixed number of neighbors regardless of the local statistics around them. We proposed a new SpectralNet similarity metric based on random projection trees (rpTrees). Our experiments revealed that SpectralNet produces better clustering accuracy using rpTree similarity metric compared to $k$-nn graph with a distance metric. Also, we found out that rpTree parameters do not affect the clustering accuracy. These parameters include the leaf size and the selection of projection direction. It is computationally efficient to keep the leaf size in order of $\log(n)$, and project the points onto a random direction instead of trying to find the direction with the maximum dispersion.
A parameter-free graph reduction for spectral clustering and SpectralNet
Feb 25, 2023Abstract:Graph-based clustering methods like spectral clustering and SpectralNet are very efficient in detecting clusters of non-convex shapes. Unlike the popular $k$-means, graph-based clustering methods do not assume that each cluster has a single mean. However, these methods need a graph where vertices in the same cluster are connected by edges of large weights. To achieve this goal, many studies have proposed graph reduction methods with parameters. Unfortunately, these parameters have to be tuned for every dataset. We introduce a graph reduction method that does not require any parameters. First, the distances from every point $p$ to its neighbors are filtered using an adaptive threshold to only keep neighbors with similar surrounding density. Second, the similarities with close neighbors are computed and only high similarities are kept. The edges that survive these two filtering steps form the constructed graph that was passed to spectral clustering and SpectralNet. The experiments showed that our method provides a stable alternative, where other methods performance fluctuated according to the setting of their parameters.
The Effect of Points Dispersion on the $k$-nn Search in Random Projection Forests
Feb 25, 2023Abstract:Partitioning trees are efficient data structures for $k$-nearest neighbor search. Machine learning libraries commonly use a special type of partitioning trees called $k$d-trees to perform $k$-nn search. Unfortunately, $k$d-trees can be ineffective in high dimensions because they need more tree levels to decrease the vector quantization (VQ) error. Random projection trees rpTrees solve this scalability problem by using random directions to split the data. A collection of rpTrees is called rpForest. $k$-nn search in an rpForest is influenced by two factors: 1) the dispersion of points along the random direction and 2) the number of rpTrees in the rpForest. In this study, we investigate how these two factors affect the $k$-nn search with varying $k$ values and different datasets. We found that with larger number of trees, the dispersion of points has a very limited effect on the $k$-nn search. One should use the original rpTree algorithm by picking a random direction regardless of the dispersion of points.
Random Projection Forest Initialization for Graph Convolutional Networks
Feb 22, 2023Abstract:Graph convolutional networks (GCNs) were a great step towards extending deep learning to unstructured data such as graphs. But GCNs still need a constructed graph to work with. To solve this problem, classical graphs such as $k$-nearest neighbor are usually used to initialize the GCN. Although it is computationally efficient to construct $k$-nn graphs, the constructed graph might not be very useful for learning. In a $k$-nn graph, points are restricted to have a fixed number of edges, and all edges in the graph have equal weights. We present a new way to construct the graph and initialize the GCN. It is based on random projection forest (rpForest). rpForest enables us to assign varying weights on edges indicating varying importance, which enhanced the learning. The number of trees is a hyperparameter in rpForest. We performed spectral analysis to help us setting this parameter in the right range. In the experiments, initializing the GCN using rpForest provides better results compared to $k$-nn initialization.
Approximate spectral clustering density-based similarity for noisy datasets
Feb 22, 2023Abstract:Approximate spectral clustering (ASC) was developed to overcome heavy computational demands of spectral clustering (SC). It maintains SC ability in predicting non-convex clusters. Since it involves a preprocessing step, ASC defines new similarity measures to assign weights on graph edges. Connectivity matrix (CONN) is an efficient similarity measure to construct graphs for ASC. It defines the weight between two vertices as the number of points assigned to them during vector quantization training. However, this relationship is undirected, where it is not clear which of the vertices is contributing more to that edge. Also, CONN could be tricked by noisy density between clusters. We defined a directed version of CONN, named DCONN, to get insights on vertices contributions to edges. Also, we provided filtering schemes to ensure CONN edges are highlighting potential clusters. Experiments reveal that the proposed filtering was highly efficient when noise cannot be tolerated by CONN.
Approximate spectral clustering with eigenvector selection and self-tuned $k$
Feb 22, 2023Abstract:The recently emerged spectral clustering surpasses conventional clustering methods by detecting clusters of any shape without the convexity assumption. Unfortunately, with a computational complexity of $O(n^3)$, it was infeasible for multiple real applications, where $n$ could be large. This stimulates researchers to propose the approximate spectral clustering (ASC). However, most of ASC methods assumed that the number of clusters $k$ was known. In practice, manual setting of $k$ could be subjective or time consuming. The proposed algorithm has two relevance metrics for estimating $k$ in two vital steps of ASC. One for selecting the eigenvectors spanning the embedding space, and the other to discover the number of clusters in that space. The algorithm used a growing neural gas (GNG) approximation, GNG is superior in preserving input data topology. The experimental setup demonstrates the efficiency of the proposed algorithm and its ability to compete with similar methods where $k$ was set manually.
Refining a $k$-nearest neighbor graph for a computationally efficient spectral clustering
Feb 22, 2023



Abstract:Spectral clustering became a popular choice for data clustering for its ability of uncovering clusters of different shapes. However, it is not always preferable over other clustering methods due to its computational demands. One of the effective ways to bypass these computational demands is to perform spectral clustering on a subset of points (data representatives) then generalize the clustering outcome, this is known as approximate spectral clustering (ASC). ASC uses sampling or quantization to select data representatives. This makes it vulnerable to 1) performance inconsistency (since these methods have a random step either in initialization or training), 2) local statistics loss (because the pairwise similarities are extracted from data representatives instead of data points). We proposed a refined version of $k$-nearest neighbor graph, in which we keep data points and aggressively reduce number of edges for computational efficiency. Local statistics were exploited to keep the edges that do not violate the intra-cluster distances and nullify all other edges in the $k$-nearest neighbor graph. We also introduced an optional step to automatically select the number of clusters $C$. The proposed method was tested on synthetic and real datasets. Compared to ASC methods, the proposed method delivered a consistent performance despite significant reduction of edges.
A Visual Measure of Changes to Weighted Self-Organizing Map Patterns
Mar 27, 2017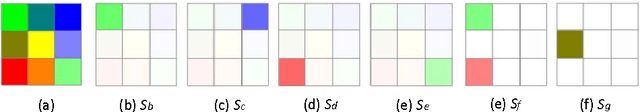



Abstract:Estimating output changes by input changes is the main task in causal analysis. In previous work, input and output Self-Organizing Maps (SOMs) were associated for causal analysis of multivariate and nonlinear data. Based on the association, a weight distribution of the output conditional on a given input was obtained over the output map space. Such a weighted SOM pattern of the output changes when the input changes. In order to analyze the change, it is important to measure the difference of the patterns. Many methods have been proposed for the dissimilarity measure of patterns. However, it remains a major challenge when attempting to measure how the patterns change. In this paper, we propose a visualization approach that simplifies the comparison of the difference in terms of the pattern property. Using this approach, the change can be analyzed by integrating colors and star glyph shapes representing the property dissimilarity. Ecological data is used to demonstrate the usefulness of our approach and the experimental results show that our approach provides the change information effectively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge