Marianna Milano
Learning Weighted Association Rules in Human Phenotype Ontology
Dec 31, 2016

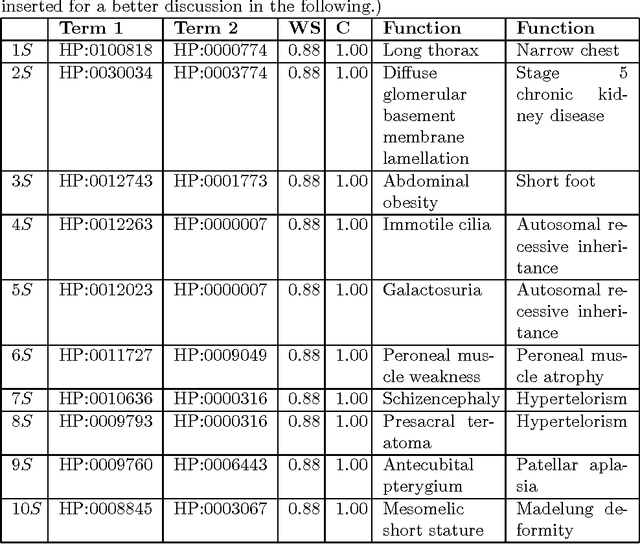

Abstract:The Human Phenotype Ontology (HPO) is a structured repository of concepts (HPO Terms) that are associated to one or more diseases. The process of association is referred to as annotation. The relevance and the specificity of both HPO terms and annotations are evaluated by a measure defined as Information Content (IC). The analysis of annotated data is thus an important challenge for bioinformatics. There exist different approaches of analysis. From those, the use of Association Rules (AR) may provide useful knowledge, and it has been used in some applications, e.g. improving the quality of annotations. Nevertheless classical association rules algorithms do not take into account the source of annotation nor the importance yielding to the generation of candidate rules with low IC. This paper presents HPO-Miner (Human Phenotype Ontology-based Weighted Association Rules) a methodology for extracting Weighted Association Rules. HPO-Miner can extract relevant rules from a biological point of view. A case study on using of HPO-Miner on publicly available HPO annotation datasets is used to demonstrate the effectiveness of our methodology.
A web-based tool to Analyze Semantic Similarity Networks
Dec 21, 2014
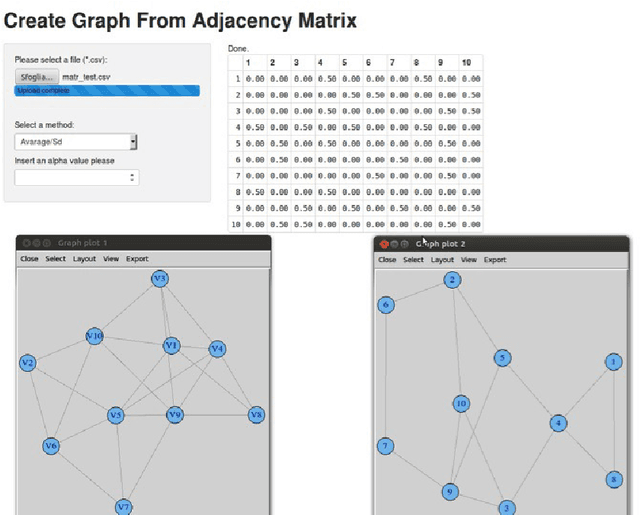

Abstract:In computational biology, biological entities such as genes or proteins are usually annotated with terms extracted from Gene Ontology (GO). The functional similarity among terms of an ontology is evaluated by using Semantic Similarity Measures (SSM). More recently, the extensive application of SSMs yielded to the Semantic Similarity Networks (SSNs). SSNs are edge-weighted graphs where the nodes are concepts (e.g. proteins) and each edge has an associated weight that represents the semantic similarity among related pairs of nodes. The analysis of SSNs may reveal biologically meaningful knowledge. For these aims, the need for the introduction of tool able to manage and analyze SSN arises. Consequently we developed SSN-Analyzer a web based tool able to build and preprocess SSN. As proof of concept we demonstrate that community detection algorithms applied to filtered (thresholded) networks, have better performances in terms of biological relevance of the results, with respect to the use of raw unfiltered networks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge