Majid Razmara
Out-of-Distribution Detection for Dermoscopic Image Classification
Apr 19, 2021
Abstract:Medical image diagnosis can be achieved by deep neural networks, provided there is enough varied training data for each disease class. However, a hitherto unknown disease class not encountered during training will inevitably be misclassified, even if predicted with low probability. This problem is especially important for medical image diagnosis, when an image of a hitherto unknown disease is presented for diagnosis, especially when the images come from the same image domain, such as dermoscopic skin images. Current out-of-distribution detection algorithms act unfairly when the in-distribution classes are imbalanced, by favouring the most numerous disease in the training sets. This could lead to false diagnoses for rare cases which are often medically important. We developed a novel yet simple method to train neural networks, which enables them to classify in-distribution dermoscopic skin disease images and also detect novel diseases from dermoscopic images at test time. We show that our BinaryHeads model not only does not hurt classification balanced accuracy when the data is imbalanced, but also consistently improves the balanced accuracy. We also introduce an important method to investigate the effectiveness of out-of-distribution detection methods based on presence of varying amounts of out-of-distribution data, which may arise in real-world settings.
Diagnostic Accuracy of Content Based Dermatoscopic Image Retrieval with Deep Classification Features
Oct 22, 2018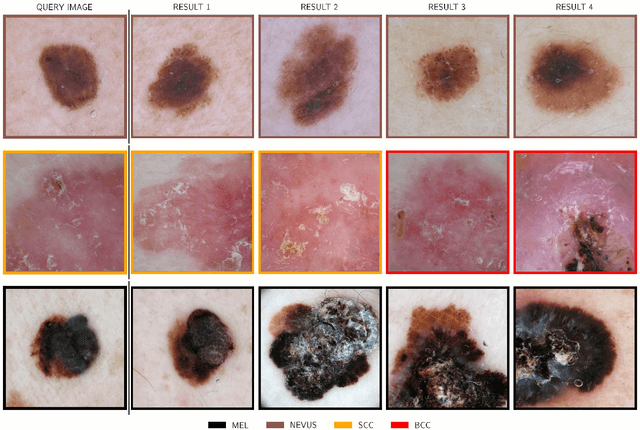
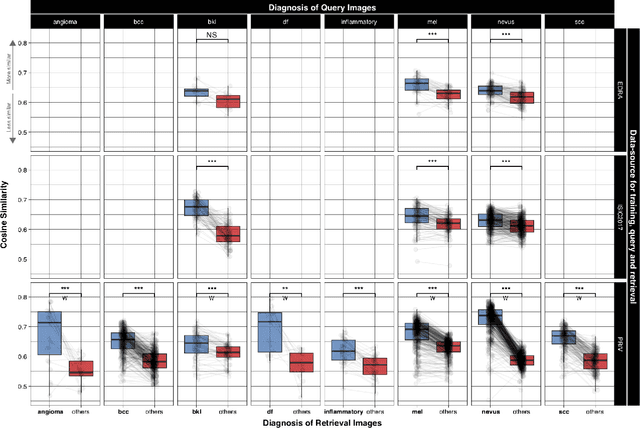
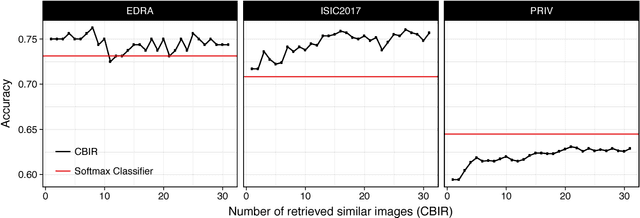
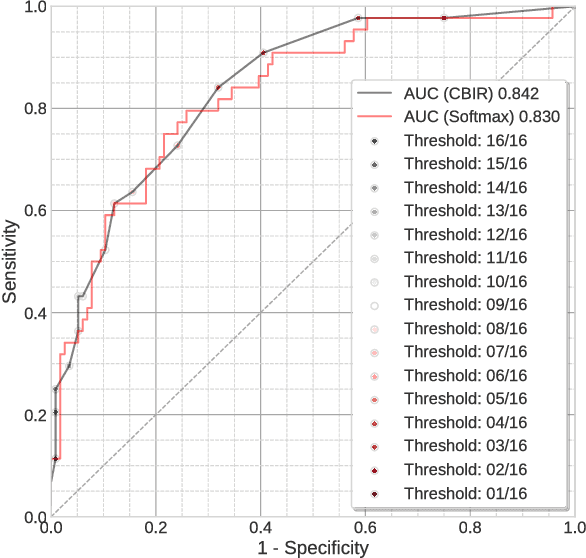
Abstract:Background: Automated classification of medical images through neural networks can reach high accuracy rates but lack interpretability. Objectives: To compare the diagnostic accuracy obtained by using content based image retrieval (CBIR) to retrieve visually similar dermatoscopic images with corresponding disease labels against predictions made by a neural network. Methods: A neural network was trained to predict disease classes on dermatoscopic images from three retrospectively collected image datasets containing 888, 2750 and 16691 images respectively. Diagnosis predictions were made based on the most commonly occurring diagnosis in visually similar images, or based on the top-1 class prediction of the softmax output from the network. Outcome measures were area under the ROC curve for predicting a malignant lesion (AUC), multiclass-accuracy and mean average precision (mAP), measured on unseen test images of the corresponding dataset. Results: In all three datasets the skin cancer predictions from CBIR (evaluating the 16 most similar images) showed AUC values similar to softmax predictions (0.842, 0.806 and 0.852 versus 0.830, 0.810 and 0.847 respectively; p-value>0.99 for all). Similarly, the multiclass-accuracy of CBIR was comparable to softmax predictions. Networks trained for detecting only 3 classes performed better on a dataset with 8 classes when using CBIR as compared to softmax predictions (mAP 0.184 vs. 0.368 and 0.198 vs. 0.403 respectively). Conclusions: Presenting visually similar images based on features from a neural network shows comparable accuracy to the softmax probability-based diagnoses of convolutional neural networks. CBIR may be more helpful than a softmax classifier in improving diagnostic accuracy of clinicians in a routine clinical setting.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge