Kristi Bushman
A Simplified Approach to Deep Learning for Image Segmentation
Aug 31, 2018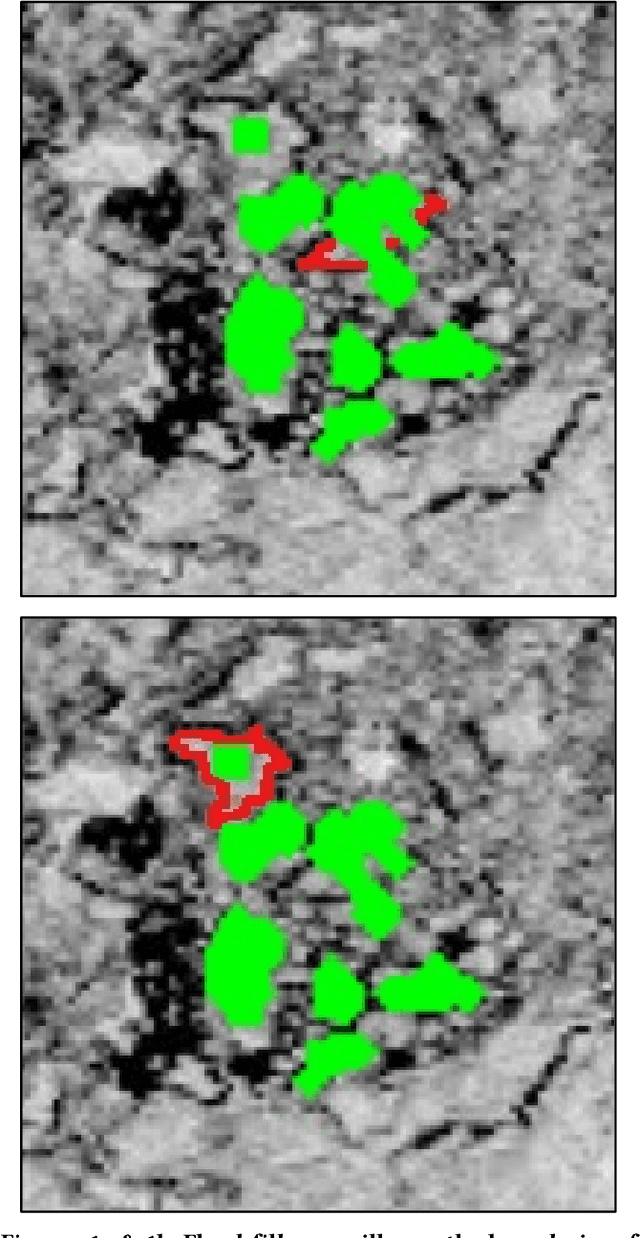



Abstract:Leaping into the rapidly developing world of deep learning is an exciting and sometimes confusing adventure. All of the advice and tutorials available can be hard to organize and work through, especially when training specific models on specific datasets, different from those originally used to train the network. In this short guide, we aim to walk the reader through the techniques that we have used to successfully train two deep neural networks for pixel-wise classification, including some data management and augmentation approaches for working with image data that may be insufficiently annotated or relatively homogenous.
* 8 pages, 6 figures (1a to 6c, plus 5 in appendix), PEARC '18: Practice and Experience in Advanced Research Computing, July 22--26, 2018, Pittsburgh, PA, USA
Understanding Neural Pathways in Zebrafish through Deep Learning and High Resolution Electron Microscope Data
Aug 31, 2018



Abstract:The tracing of neural pathways through large volumes of image data is an incredibly tedious and time-consuming process that significantly encumbers progress in neuroscience. We are exploring deep learning's potential to automate segmentation of high-resolution scanning electron microscope (SEM) image data to remove that barrier. We have started with neural pathway tracing through 5.1GB of whole-brain serial-section slices from larval zebrafish collected by the Center for Brain Science at Harvard University. This kind of manual image segmentation requires years of careful work to properly trace the neural pathways in an organism as small as a zebrafish larva (approximately 5mm in total body length). In automating this process, we would vastly improve productivity, leading to faster data analysis and breakthroughs in understanding the complexity of the brain. We will build upon prior attempts to employ deep learning for automatic image segmentation extending methods for unconventional deep learning data.
* 8 pages, 5 figures (1a to 5c), PEARC '18: Practice and Experience in Advanced Research Computing, July 22--26, 2018, Pittsburgh, PA, USA
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge