Kevin Raina
Logit Disagreement: OoD Detection with Bayesian Neural Networks
Feb 21, 2025Abstract:Bayesian neural networks (BNNs), which estimate the full posterior distribution over model parameters, are well-known for their role in uncertainty quantification and its promising application in out-of-distribution detection (OoD). Amongst other uncertainty measures, BNNs provide a state-of-the art estimation of predictive entropy (total uncertainty) which can be decomposed as the sum of mutual information and expected entropy. In the context of OoD detection the estimation of predictive uncertainty in the form of the predictive entropy score confounds aleatoric and epistemic uncertainty, the latter being hypothesized to be high for OoD points. Despite these justifications, the mutual information score has been shown to perform worse than predictive entropy. Taking inspiration from Bayesian variational autoencoder (BVAE) literature, this work proposes to measure the disagreement between a corrected version of the pre-softmax quantities, otherwise known as logits, as an estimate of epistemic uncertainty for Bayesian NNs under mean field variational inference. The three proposed epistemic uncertainty scores demonstrate marked improvements over mutual information on a range of OoD experiments, with equal performance otherwise. Moreover, the epistemic uncertainty scores perform on par with the Bayesian benchmark predictive entropy on a range of MNIST and CIFAR10 experiments.
Statistical inference of the inter-sample Dice distribution for discriminative CNN brain lesion segmentation models
Dec 04, 2020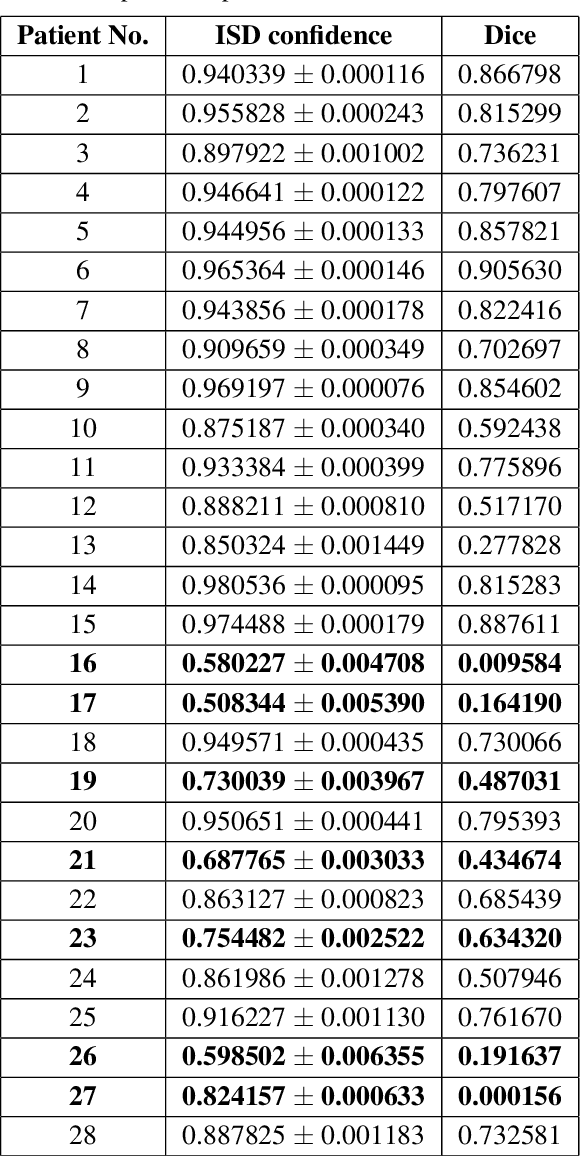
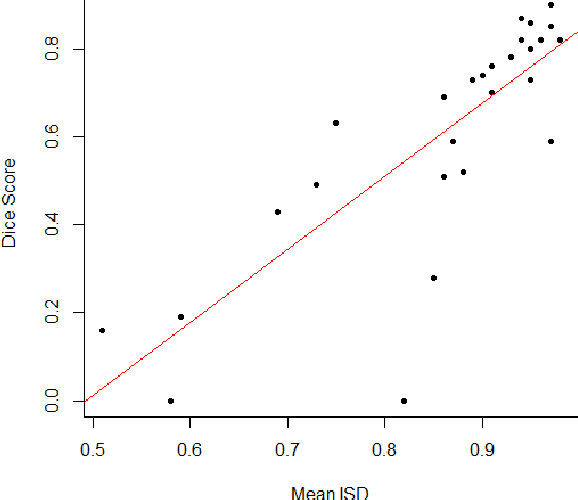
Abstract:Discriminative convolutional neural networks (CNNs), for which a voxel-wise conditional Multinoulli distribution is assumed, have performed well in many brain lesion segmentation tasks. For a trained discriminative CNN to be used in clinical practice, the patient's radiological features are inputted into the model, in which case a conditional distribution of segmentations is produced. Capturing the uncertainty of the predictions can be useful in deciding whether to abandon a model, or choose amongst competing models. In practice, however, we never know the ground truth segmentation, and therefore can never know the true model variance. In this work, segmentation sampling on discriminative CNNs is used to assess a trained model's robustness by analyzing the inter-sample dice distribution on a new patient solely based on their magnetic resonance (MR) images. Furthermore, by demonstrating the inter-sample Dice observations are independent and identically distributed with a finite mean and variance under certain conditions, a rigorous confidence based decision rule is proposed to decide whether to reject or accept a CNN model for a particular patient. Applied to the ISLES 2015 (SISS) dataset, the model identified 7 predictions as non-robust, and the average Dice coefficient calculated on the remaining brains improved by 12 percent.
Modelling brain lesion volume in patches with CNN-based Poisson Regression
Nov 26, 2020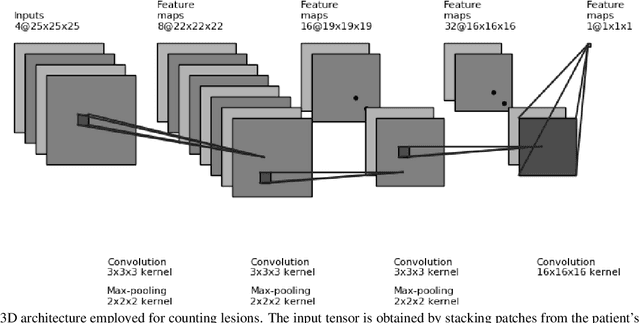
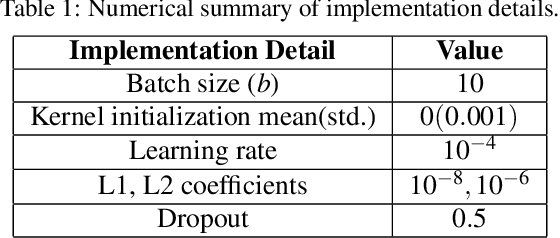
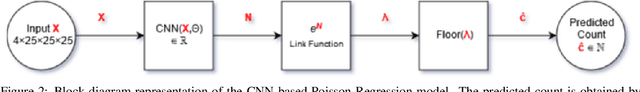
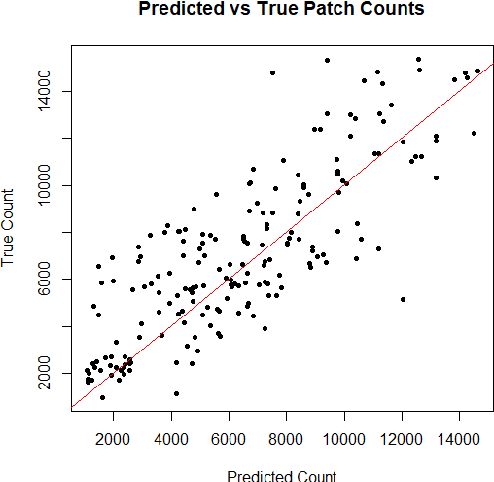
Abstract:Monitoring the progression of lesions is important for clinical response. Summary statistics such as lesion volume are objective and easy to interpret, which can help clinicians assess lesion growth or decay. CNNs are commonly used in medical image segmentation for their ability to produce useful features within large contexts and their associated efficient iterative patch-based training. Many CNN architectures require hundreds of thousands parameters to yield a good segmentation. In this work, an efficient, computationally inexpensive CNN is implemented to estimate the number of lesion voxels in a predefined patch size from magnetic resonance (MR) images. The output of the CNN is interpreted as the conditional Poisson parameter over the patch, allowing standard mini-batch gradient descent to be employed. The ISLES2015 (SISS) data is used to train and evaluate the model, which by estimating lesion volume from raw features, accurately identified the lesion image with the larger lesion volume for 86% of paired sample patches. An argument for the development and use of estimating lesion volumes to also aid in model selection for segmentation is made.
Exploiting bilateral symmetry in brain lesion segmentation
Jul 18, 2019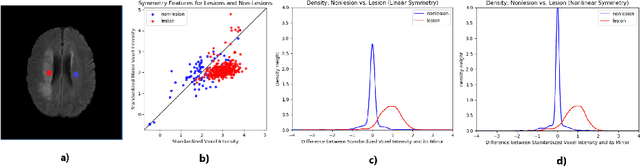
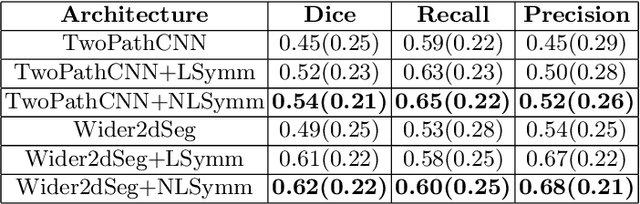
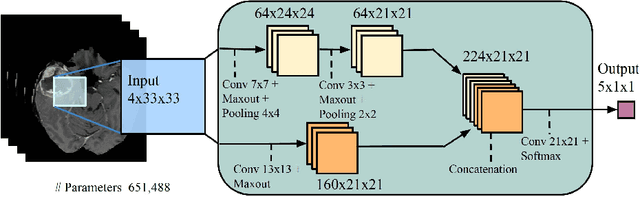
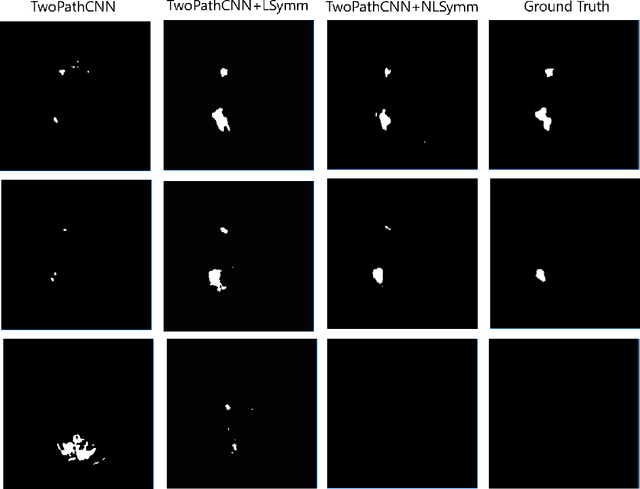
Abstract:Brain lesions, including stroke and tumours, have a high degree of variability in terms of location, size, intensity and form, making automatic segmentation difficult. We propose an improvement to existing segmentation methods by exploiting the bilateral quasi-symmetry of healthy brains, which breaks down when lesions are present. Specifically, we use nonlinear registration of a neuroimage to a reflected version of itself ("reflective registration") to determine for each voxel its homologous (corresponding) voxel in the other hemisphere. A patch around the homologous voxel is added as a set of new features to the segmentation algorithm. To evaluate this method, we implemented two different CNN-based multimodal MRI stroke lesion segmentation algorithms, and then augmented them by adding extra symmetry features using the reflective registration method described above. For each architecture, we compared the performance with and without symmetry augmentation, on the SISS Training dataset of the Ischemic Stroke Lesion Segmentation Challenge (ISLES) 2015 challenge. Using affine reflective registration improves performance over baseline, but nonlinear reflective registration gives significantly better results: an improvement in Dice coefficient of 13 percentage points over baseline for one architecture and 9 points for the other. We argue for the broad applicability of adding symmetric features to existing segmentation algorithms, specifically using nonlinear, template-free methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge