Jhony-Heriberto Giraldo-Zuluaga
Camera-trap images segmentation using multi-layer robust principal component analysis
Dec 30, 2017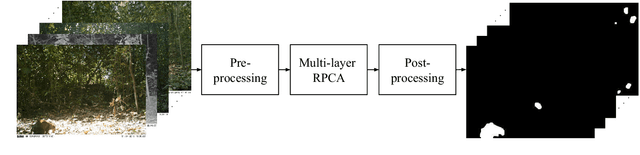
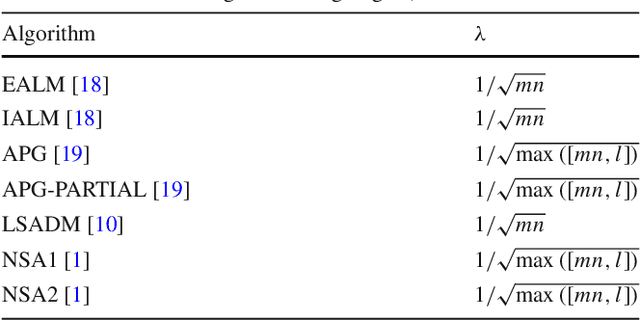
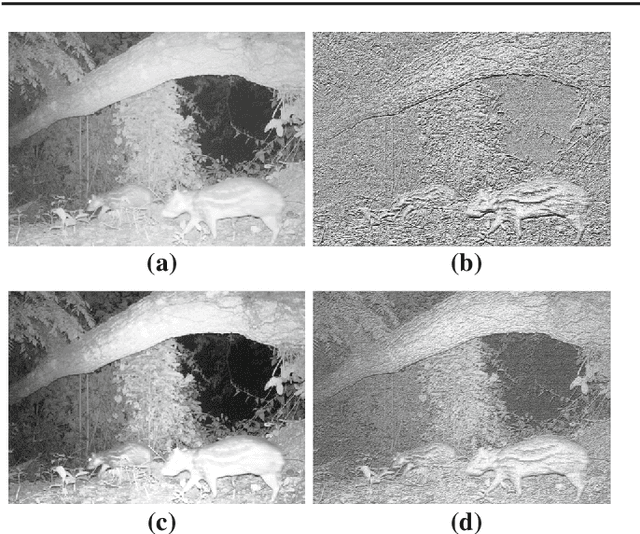
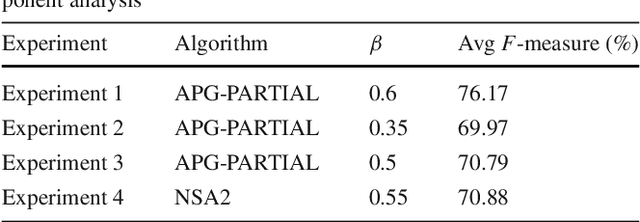
Abstract:The segmentation of animals from camera-trap images is a difficult task. To illustrate, there are various challenges due to environmental conditions and hardware limitation in these images. We proposed a multi-layer robust principal component analysis (multi-layer RPCA) approach for background subtraction. Our method computes sparse and low-rank images from a weighted sum of descriptors, using color and texture features as case of study for camera-trap images segmentation. The segmentation algorithm is composed of histogram equalization or Gaussian filtering as pre-processing, and morphological filters with active contour as post-processing. The parameters of our multi-layer RPCA were optimized with an exhaustive search. The database consists of camera-trap images from the Colombian forest taken by the Instituto de Investigaci\'on de Recursos Biol\'ogicos Alexander von Humboldt. We analyzed the performance of our method in inherent and therefore challenging situations of camera-trap images. Furthermore, we compared our method with some state-of-the-art algorithms of background subtraction, where our multi-layer RPCA outperformed these other methods. Our multi-layer RPCA reached 76.17 and 69.97% of average fine-grained F-measure for color and infrared sequences, respectively. To our best knowledge, this paper is the first work proposing multi-layer RPCA and using it for camera-trap images segmentation.
* This is a pre-print of an article published in The Visual Computer. The final authenticated version is available online at: https://doi.org/10.1007/s00371-017-1463-9
Automatic Identification of Scenedesmus Polymorphic Microalgae from Microscopic Images
Oct 23, 2017
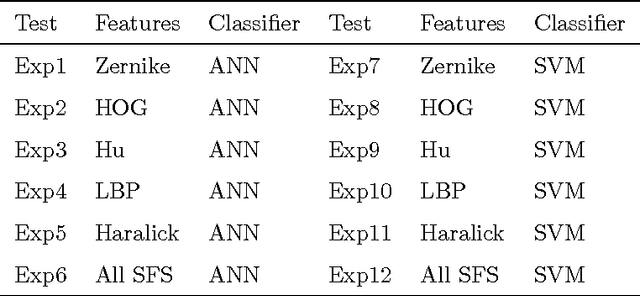

Abstract:Microalgae counting is used to measure biomass quantity. Usually, it is performed in a manual way using a Neubauer chamber and expert criterion, with the risk of a high error rate. This paper addresses the methodology for automatic identification of Scenedesmus microalgae (used in the methane production and food industry) and applies it to images captured by a digital microscope. The use of contrast adaptive histogram equalization for pre-processing, and active contours for segmentation are presented. The calculation of statistical features (Histogram of Oriented Gradients, Hu and Zernike moments) with texture features (Haralick and Local Binary Patterns descriptors) are proposed for algae characterization. Scenedesmus algae can build coenobia consisting of 1, 2, 4 and 8 cells. The amount of algae of each coenobium helps to determine the amount of lipids, proteins, and other substances in a given sample of a algae crop. The knowledge of the quantity of those elements improves the quality of bioprocess applications. Classification of coenobia achieves accuracies of 98.63% and 97.32% with Support Vector Machine (SVM) and Artificial Neural Network (ANN), respectively. According to the results it is possible to consider the proposed methodology as an alternative to the traditional technique for algae counting. The database used in this paper is publicly available for download.
Automatic Recognition of Mammal Genera on Camera-Trap Images using Multi-Layer Robust Principal Component Analysis and Mixture Neural Networks
May 08, 2017


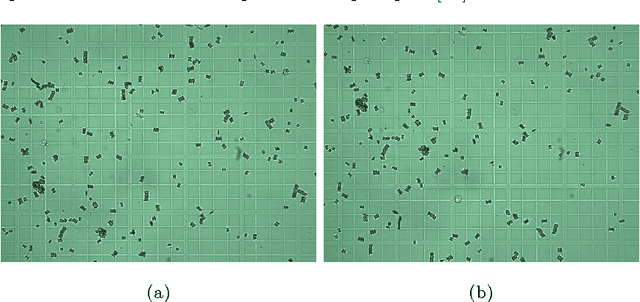
Abstract:The segmentation and classification of animals from camera-trap images is due to the conditions under which the images are taken, a difficult task. This work presents a method for classifying and segmenting mammal genera from camera-trap images. Our method uses Multi-Layer Robust Principal Component Analysis (RPCA) for segmenting, Convolutional Neural Networks (CNNs) for extracting features, Least Absolute Shrinkage and Selection Operator (LASSO) for selecting features, and Artificial Neural Networks (ANNs) or Support Vector Machines (SVM) for classifying mammal genera present in the Colombian forest. We evaluated our method with the camera-trap images from the Alexander von Humboldt Biological Resources Research Institute. We obtained an accuracy of 92.65% classifying 8 mammal genera and a False Positive (FP) class, using automatic-segmented images. On the other hand, we reached 90.32% of accuracy classifying 10 mammal genera, using ground-truth images only. Unlike almost all previous works, we confront the animal segmentation and genera classification in the camera-trap recognition. This method shows a new approach toward a fully-automatic detection of animals from camera-trap images.
Semi-Supervised Recognition of the Diploglossus Millepunctatus Lizard Species using Artificial Vision Algorithms
Nov 09, 2016



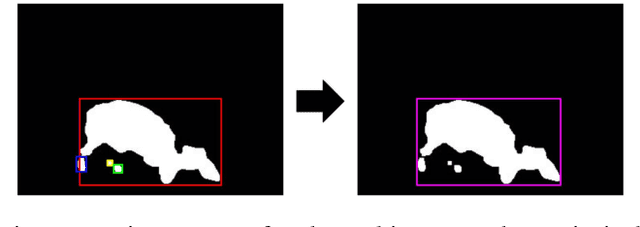
Abstract:Animal biometrics is an important requirement for monitoring and conservation tasks. The classical animal biometrics risk the animals' integrity, are expensive for numerous animals, and depend on expert criterion. The non-invasive biometrics techniques offer alternatives to manage the aforementioned problems. In this paper we propose an automatic segmentation and identification algorithm based on artificial vision algorithms to recognize Diploglossus millepunctatus. Diploglossus millepunctatus is an endangered lizard species. The algorithm is based on two stages: automatic segmentation to remove the subjective evaluation, and one identification stage to reduce the analysis time. A 82.87% of correct segmentation in average is reached. Meanwhile the identification algorithm is achieved with euclidean distance point algorithms such as Iterative Closest Point and Procrustes Analysis. A performance of 92.99% on the top 1, and a 96.82% on the top 5 is reached. The developed software, and the database used in this paper are publicly available for download from the web page of the project.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge