Henry Marichal
DeepCS-TRD, a Deep Learning-based Cross-Section Tree Ring Detector
Apr 22, 2025Abstract:Here, we propose Deep CS-TRD, a new automatic algorithm for detecting tree rings in whole cross-sections. It substitutes the edge detection step of CS-TRD by a deep-learning-based approach (U-Net), which allows the application of the method to different image domains: microscopy, scanner or smartphone acquired, and species (Pinus taeda, Gleditsia triachantos and Salix glauca). Additionally, we introduce two publicly available datasets of annotated images to the community. The proposed method outperforms state-of-the-art approaches in macro images (Pinus taeda and Gleditsia triacanthos) while showing slightly lower performance in microscopy images of Salix glauca. To our knowledge, this is the first paper that studies automatic tree ring detection for such different species and acquisition conditions. The dataset and source code are available in https://github.com/hmarichal93/deepcstrd
A Brief Analysis of the Iterative Next Boundary Detection Network for Tree Rings Delineation in Images of Pinus taeda
Aug 26, 2024Abstract:This work presents the INBD network proposed by Gillert et al. in CVPR-2023 and studies its application for delineating tree rings in RGB images of Pinus taeda cross sections captured by a smartphone (UruDendro dataset), which are images with different characteristics from the ones used to train the method. The INBD network operates in two stages: first, it segments the background, pith, and ring boundaries. In the second stage, the image is transformed into polar coordinates, and ring boundaries are iteratively segmented from the pith to the bark. Both stages are based on the U-Net architecture. The method achieves an F-Score of 77.5, a mAR of 0.540, and an ARAND of 0.205 on the evaluation set. The code for the experiments is available at https://github.com/hmarichal93/mlbrief_inbd.
UruDendro, a public dataset of cross-section images of Pinus taeda
Apr 16, 2024

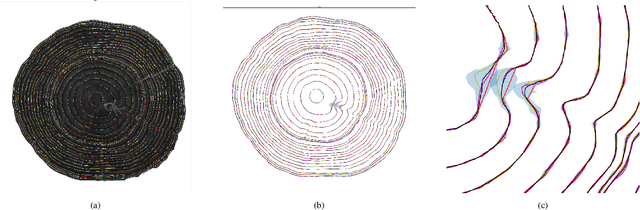

Abstract:The automatic detection of tree-ring boundaries and other anatomical features using image analysis has progressed substantially over the past decade with advances in machine learning and imagery technology, as well as increasing demands from the dendrochronology community. This paper presents a publicly available database of 64 scanned images of transverse sections of commercially grown Pinus taeda trees from northern Uruguay, ranging from 17 to 24 years old. The collection contains several challenging features for automatic ring detection, including illumination and surface preparation variation, fungal infection (blue stains), knot formation, missing cortex or interruptions in outer rings, and radial cracking. This dataset can be used to develop and test automatic tree ring detection algorithms. This paper presents to the dendrochronology community one such method, Cross-Section Tree-Ring Detection (CS-TRD), which identifies and marks complete annual rings in cross-sections for tree species presenting a clear definition between early and latewood. We compare the CS-TRD performance against the ground truth manual delineation of all rings over the UruDendro dataset. The CS-TRD software identified rings with an average F-score of 89% and RMSE error of 5.27px for the entire database in less than 20 seconds per image. Finally, we propose a robust measure of the ring growth using the \emph{equivalent radius} of a circle having the same area enclosed by the detected tree ring. Overall, this study contributes to the dendrochronologist's toolbox of fast and low-cost methods to automatically detect rings in conifer species, particularly for measuring diameter growth rates and stem transverse area using entire cross-sections.
Automatic Wood Pith Detector: Local Orientation Estimation and Robust Accumulation
Apr 02, 2024Abstract:A fully automated technique for wood pith detection (APD), relying on the concentric shape of the structure of wood ring slices, is introduced. The method estimates the ring's local orientations using the 2D structure tensor and finds the pith position, optimizing a cost function designed for this problem. We also present a variant (APD-PCL), using the parallel coordinates space, that enhances the method's effectiveness when there are no clear tree ring patterns. Furthermore, refining previous work by Kurdthongmee, a YoloV8 net is trained for pith detection, producing a deep learning-based approach to the same problem (APD-DL). All methods were tested on seven datasets, including images captured under diverse conditions (controlled laboratory settings, sawmill, and forest) and featuring various tree species (Pinus taeda, Douglas fir, Abies alba, and Gleditsia triacanthos). All proposed approaches outperform existing state-of-the-art methods and can be used in CPU-based real-time applications. Additionally, we provide a novel dataset comprising images of gymnosperm and angiosperm species. Dataset and source code are available at http://github.com/hmarichal93/apd.
CS-TRD: a Cross Sections Tree Ring Detection method
May 18, 2023Abstract:This work describes a Tree Ring Detection method for complete Cross-Sections of trees (CS-TRD). The method is based on the detection, processing, and connection of edges corresponding to the tree's growth rings. The method depends on the parameters for the Canny Devernay edge detector ($\sigma$ and two thresholds), a resize factor, the number of rays, and the pith location. The first five parameters are fixed by default. The pith location can be marked manually or using an automatic pith detection algorithm. Besides the pith localization, the CS-TRD method is fully automated and achieves an F-Score of 89\% in the UruDendro dataset (of Pinus Taeda) with a mean execution time of 17 seconds and of 97\% in the Kennel dataset (of Abies Alba) with an average execution time 11 seconds.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge