Gaël de Lannoy
A Mask R-CNN approach to counting bacterial colony forming units in pharmaceutical development
Mar 09, 2021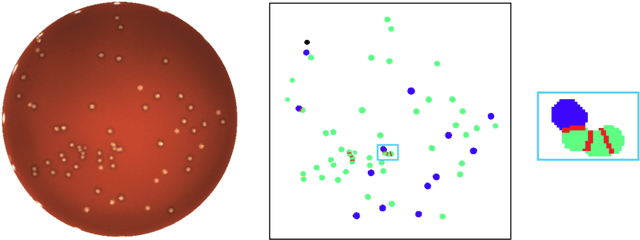
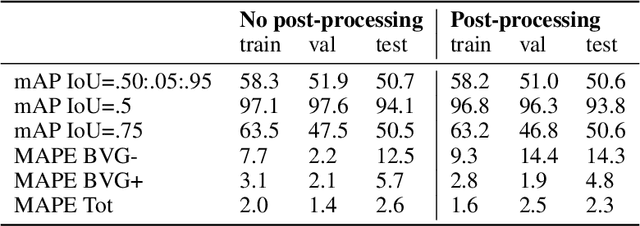
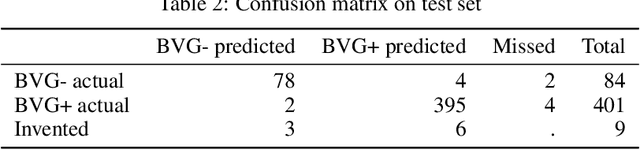
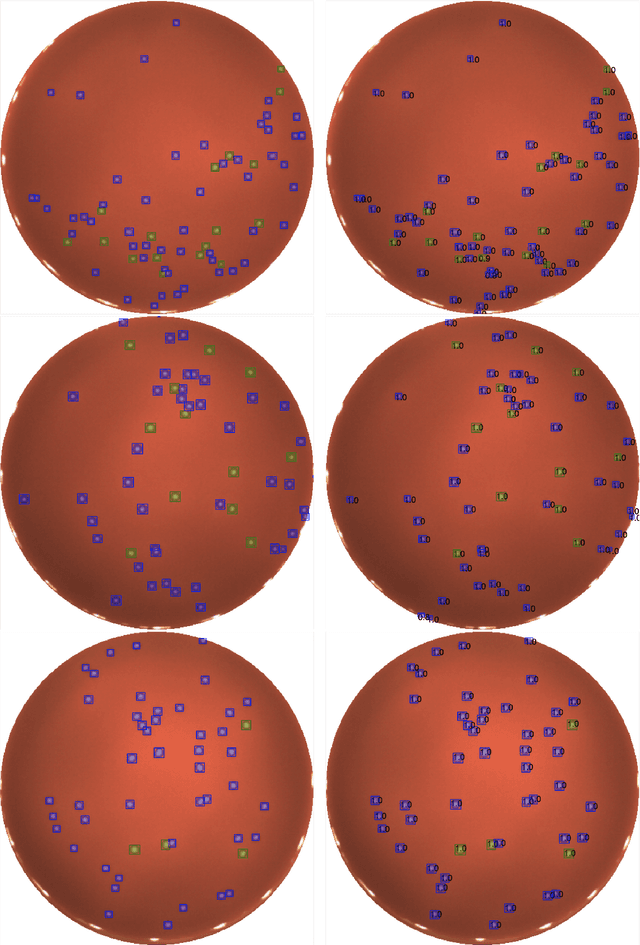
Abstract:We present an application of the well-known Mask R-CNN approach to the counting of different types of bacterial colony forming units that were cultured in Petri dishes. Our model was made available to lab technicians in a modern SPA (Single-Page Application). Users can upload images of dishes, after which the Mask R-CNN model that was trained and tuned specifically for this task detects the number of BVG- and BVG+ colonies and displays these in an interactive interface for the user to verify. Users can then check the model's predictions, correct them if deemed necessary, and finally validate them. Our adapted Mask R-CNN model achieves a mean average precision (mAP) of 94\% at an intersection-over-union (IoU) threshold of 50\%. With these encouraging results, we see opportunities to bring the benefits of improved accuracy and time saved to related problems, such as generalising to other bacteria types and viral foci counting.
Deep Learning to Detect Bacterial Colonies for the Production of Vaccines
Sep 02, 2020
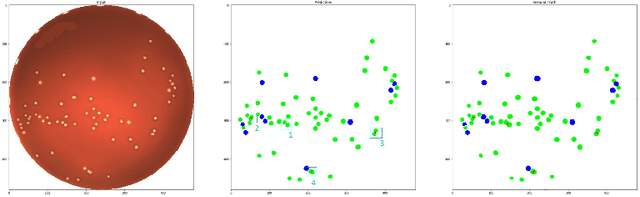
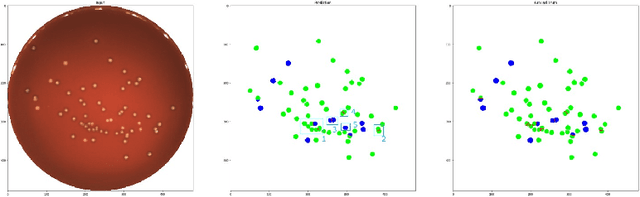
Abstract:During the development of vaccines, bacterial colony forming units (CFUs) are counted in order to quantify the yield in the fermentation process. This manual task is time-consuming and error-prone. In this work we test multiple segmentation algorithms based on the U-Net CNN architecture and show that these offer robust, automated CFU counting. We show that the multiclass generalisation with a bespoke loss function allows distinguishing virulent and avirulent colonies with acceptable accuracy. While many possibilities are left to explore, our results show the potential of deep learning for separating and classifying bacterial colonies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge