Fadeel Sher Khan
MFSR-GAN: Multi-Frame Super-Resolution with Handheld Motion Modeling
Feb 28, 2025



Abstract:Smartphone cameras have become ubiquitous imaging tools, yet their small sensors and compact optics often limit spatial resolution and introduce distortions. Combining information from multiple low-resolution (LR) frames to produce a high-resolution (HR) image has been explored to overcome the inherent limitations of smartphone cameras. Despite the promise of multi-frame super-resolution (MFSR), current approaches are hindered by datasets that fail to capture the characteristic noise and motion patterns found in real-world handheld burst images. In this work, we address this gap by introducing a novel synthetic data engine that uses multi-exposure static images to synthesize LR-HR training pairs while preserving sensor-specific noise characteristics and image motion found during handheld burst photography. We also propose MFSR-GAN: a multi-scale RAW-to-RGB network for MFSR. Compared to prior approaches, MFSR-GAN emphasizes a "base frame" throughout its architecture to mitigate artifacts. Experimental results on both synthetic and real data demonstrates that MFSR-GAN trained with our synthetic engine yields sharper, more realistic reconstructions than existing methods for real-world MFSR.
Graph-Based Biomarker Discovery and Interpretation for Alzheimer's Disease
Nov 27, 2024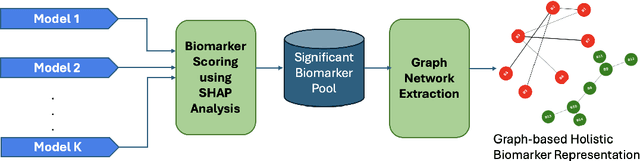
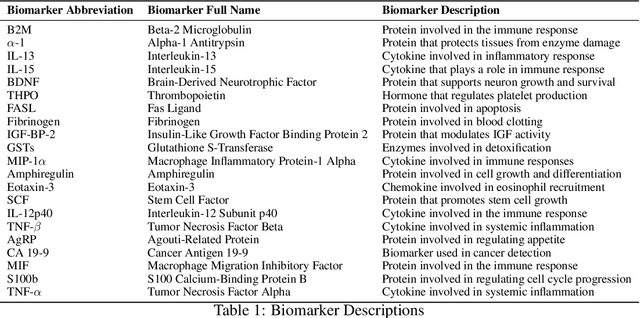
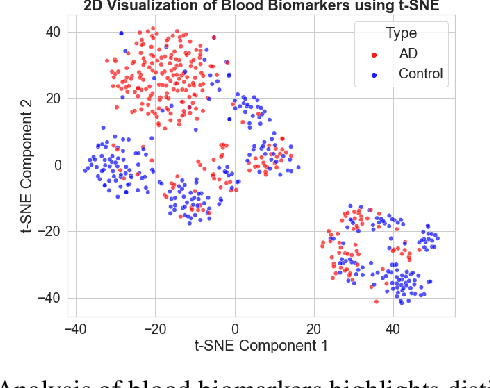
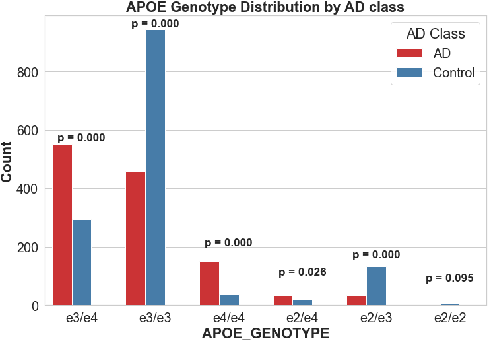
Abstract:Early diagnosis and discovery of therapeutic drug targets are crucial objectives for the effective management of Alzheimer's Disease (AD). Current approaches for AD diagnosis and treatment planning are based on radiological imaging and largely inaccessible for population-level screening due to prohibitive costs and limited availability. Recently, blood tests have shown promise in diagnosing AD and highlighting possible biomarkers that can be used as drug targets for AD management. Blood tests are significantly more accessible to disadvantaged populations, cost-effective, and minimally invasive. However, biomarker discovery in the context of AD diagnosis is complex as there exist important associations between various biomarkers. Here, we introduce BRAIN (Biomarker Representation, Analysis, and Interpretation Network), a novel machine learning (ML) framework to jointly optimize the diagnostic accuracy and biomarker discovery processes to identify all relevant biomarkers that contribute to AD diagnosis. Using a holistic graph-based representation for biomarkers, we highlight their inter-dependencies and explain why different ML models identify different discriminative biomarkers. We apply BRAIN to a publicly available blood biomarker dataset, revealing three novel biomarker sub-networks whose interactions vary between the control and AD groups, offering a new paradigm for drug discovery and biomarker analysis for AD.
Single color virtual H&E staining with In-and-Out Net
May 22, 2024



Abstract:Virtual staining streamlines traditional staining procedures by digitally generating stained images from unstained or differently stained images. While conventional staining methods involve time-consuming chemical processes, virtual staining offers an efficient and low infrastructure alternative. Leveraging microscopy-based techniques, such as confocal microscopy, researchers can expedite tissue analysis without the need for physical sectioning. However, interpreting grayscale or pseudo-color microscopic images remains a challenge for pathologists and surgeons accustomed to traditional histologically stained images. To fill this gap, various studies explore digitally simulating staining to mimic targeted histological stains. This paper introduces a novel network, In-and-Out Net, specifically designed for virtual staining tasks. Based on Generative Adversarial Networks (GAN), our model efficiently transforms Reflectance Confocal Microscopy (RCM) images into Hematoxylin and Eosin (H&E) stained images. We enhance nuclei contrast in RCM images using aluminum chloride preprocessing for skin tissues. Training the model with virtual H\&E labels featuring two fluorescence channels eliminates the need for image registration and provides pixel-level ground truth. Our contributions include proposing an optimal training strategy, conducting a comparative analysis demonstrating state-of-the-art performance, validating the model through an ablation study, and collecting perfectly matched input and ground truth images without registration. In-and-Out Net showcases promising results, offering a valuable tool for virtual staining tasks and advancing the field of histological image analysis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge