Fabián Villena
llmNER: (Zero
Jun 06, 2024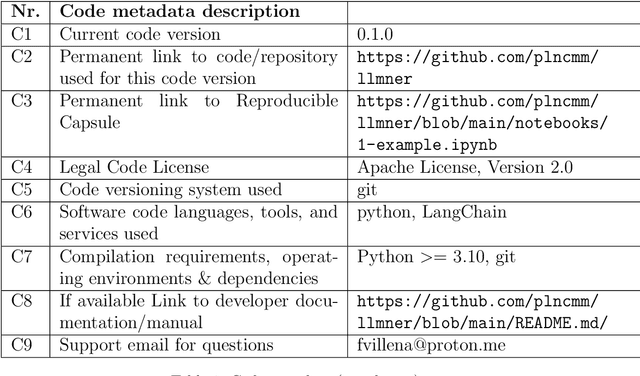
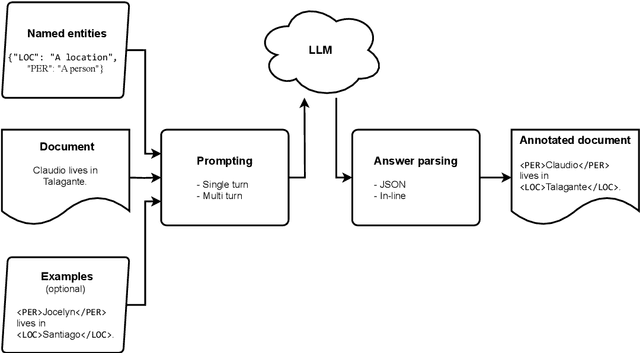
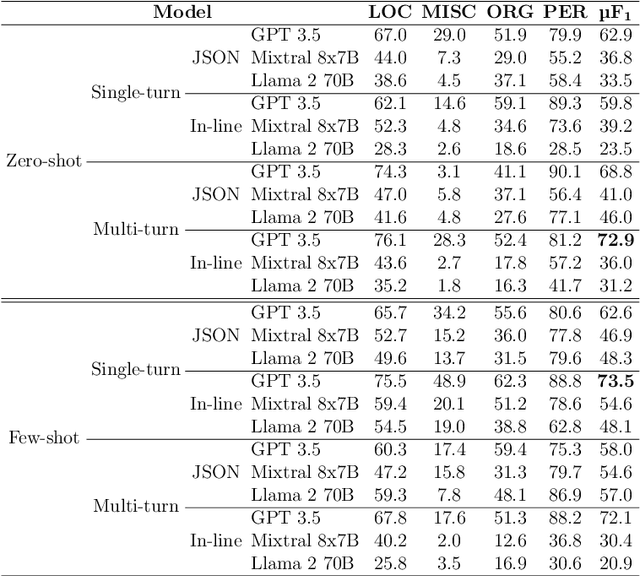
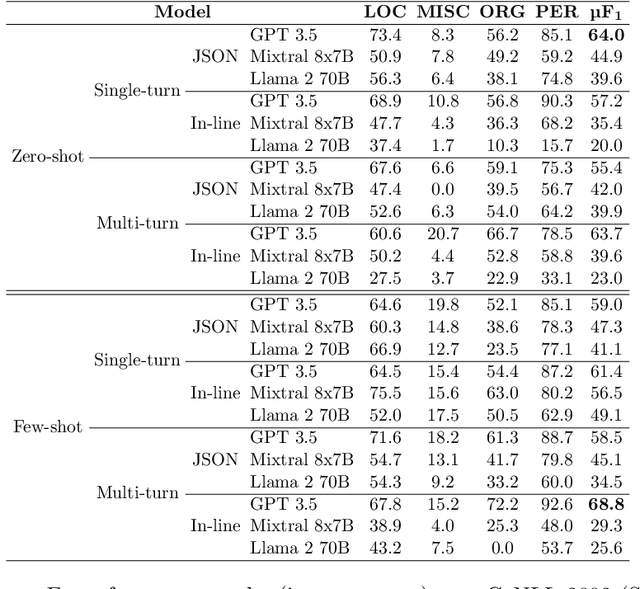
Abstract:Large language models (LLMs) allow us to generate high-quality human-like text. One interesting task in natural language processing (NLP) is named entity recognition (NER), which seeks to detect mentions of relevant information in documents. This paper presents llmNER, a Python library for implementing zero-shot and few-shot NER with LLMs; by providing an easy-to-use interface, llmNER can compose prompts, query the model, and parse the completion returned by the LLM. Also, the library enables the user to perform prompt engineering efficiently by providing a simple interface to test multiple variables. We validated our software on two NER tasks to show the library's flexibility. llmNER aims to push the boundaries of in-context learning research by removing the barrier of the prompting and parsing steps.
Automatic Coding at Scale: Design and Deployment of a Nationwide System for Normalizing Referrals in the Chilean Public Healthcare System
Jul 09, 2023Abstract:The disease coding task involves assigning a unique identifier from a controlled vocabulary to each disease mentioned in a clinical document. This task is relevant since it allows information extraction from unstructured data to perform, for example, epidemiological studies about the incidence and prevalence of diseases in a determined context. However, the manual coding process is subject to errors as it requires medical personnel to be competent in coding rules and terminology. In addition, this process consumes a lot of time and energy, which could be allocated to more clinically relevant tasks. These difficulties can be addressed by developing computational systems that automatically assign codes to diseases. In this way, we propose a two-step system for automatically coding diseases in referrals from the Chilean public healthcare system. Specifically, our model uses a state-of-the-art NER model for recognizing disease mentions and a search engine system based on Elasticsearch for assigning the most relevant codes associated with these disease mentions. The system's performance was evaluated on referrals manually coded by clinical experts. Our system obtained a MAP score of 0.63 for the subcategory level and 0.83 for the category level, close to the best-performing models in the literature. This system could be a support tool for health professionals, optimizing the coding and management process. Finally, to guarantee reproducibility, we publicly release the code of our models and experiments.
LaboRecommender: A crazy-easy to use Python-based recommender system for laboratory tests
May 03, 2021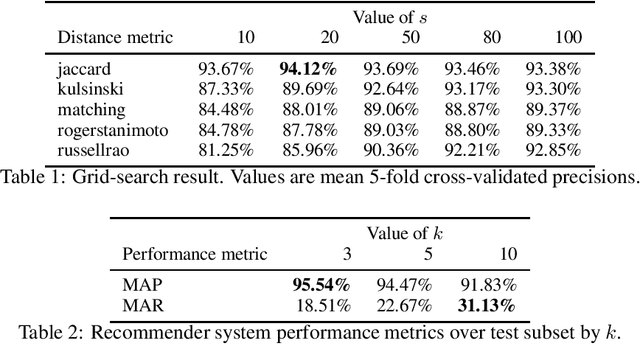
Abstract:Laboratory tests play a major role in clinical decision making because they are essential for the confirmation of diagnostics suspicions and influence medical decisions. The number of different laboratory tests available to physicians in our age has been expanding very rapidly due to the rapid advances in laboratory technology. To find the correct desired tests within this expanding plethora of elements, the Health Information System must provide a powerful search engine and the practitioner need to remember the exact name of the laboratory test to correctly select the bag of tests to order. Recommender systems are platforms which suggest appropriate items to a user after learning the users' behaviour. A neighbourhood-based collaborative filtering method was used to model the recommender system, where similar bags, clustered using nearest neighbours algorithm, are used to make recommendations of tests for each other similar bag of laboratory tests. The recommender system developed in this paper achieved 95.54 % in the mean average precision metric. A fully documented Python package named LaboRecommender was developed to implement the algorithm proposed in this paper
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge