Douglas Pinto Sampaio Gomes
Leaf Segmentation and Counting with Deep Learning: on Model Certainty, Test-Time Augmentation, Trade-Offs
Dec 21, 2020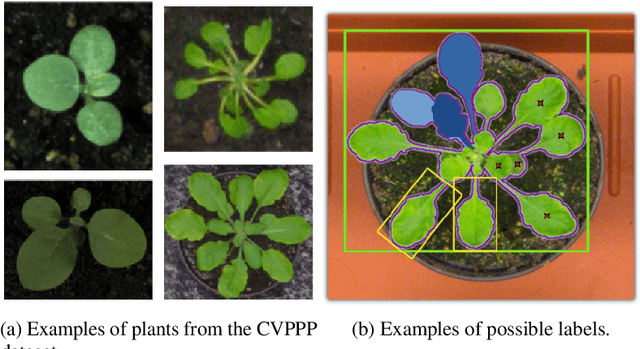
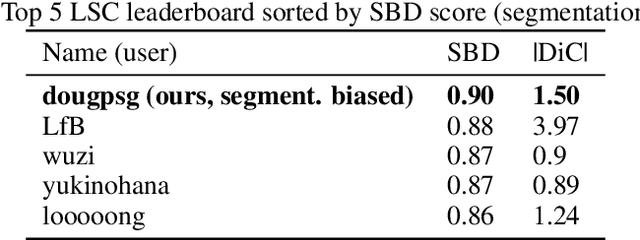


Abstract:Plant phenotyping tasks such as leaf segmentation and counting are fundamental to the study of phenotypic traits. Since it is well-suited for these tasks, deep supervised learning has been prevalent in recent works proposing better performing models at segmenting and counting leaves. Despite good efforts from research groups, one of the main challenges for proposing better methods is still the limitation of labelled data availability. The main efforts of the field seem to be augmenting existing limited data sets, and some aspects of the modelling process have been under-discussed. This paper explores such topics and present experiments that led to the development of the best-performing method in the Leaf Segmentation Challenge and in another external data set of Komatsuna plants. The model has competitive performance while been arguably simpler than other recently proposed ones. The experiments also brought insights such as the fact that model cardinality and test-time augmentation may have strong applications in object segmentation of single class and high occlusion, and regarding the data distribution of recently proposed data sets for benchmarking.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge