David Medina-Ortiz
From thermodynamics to protein design: Diffusion models for biomolecule generation towards autonomous protein engineering
Jan 05, 2025



Abstract:Protein design with desirable properties has been a significant challenge for many decades. Generative artificial intelligence is a promising approach and has achieved great success in various protein generation tasks. Notably, diffusion models stand out for their robust mathematical foundations and impressive generative capabilities, offering unique advantages in certain applications such as protein design. In this review, we first give the definition and characteristics of diffusion models and then focus on two strategies: Denoising Diffusion Probabilistic Models and Score-based Generative Models, where DDPM is the discrete form of SGM. Furthermore, we discuss their applications in protein design, peptide generation, drug discovery, and protein-ligand interaction. Finally, we outline the future perspectives of diffusion models to advance autonomous protein design and engineering. The E(3) group consists of all rotations, reflections, and translations in three-dimensions. The equivariance on the E(3) group can keep the physical stability of the frame of each amino acid as much as possible, and we reflect on how to keep the diffusion model E(3) equivariant for protein generation.
Peptipedia: a comprehensive database for peptide research supported by Assembled predictive models and Data Mining approaches
Jan 28, 2021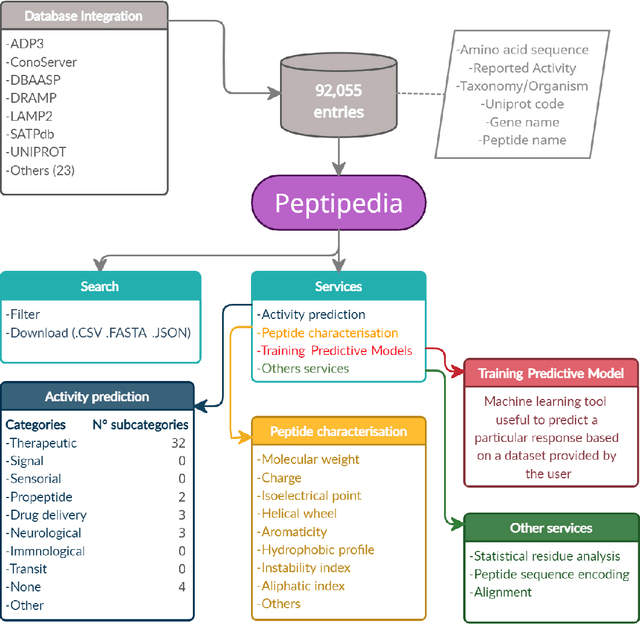
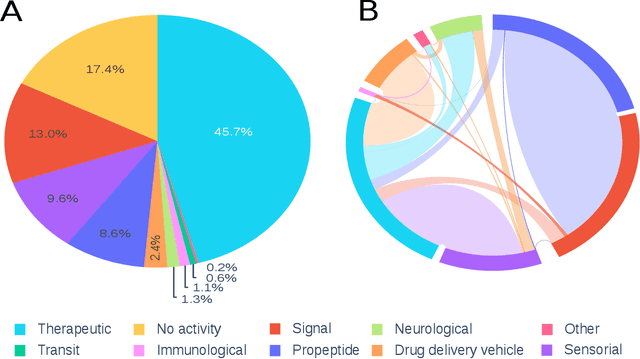
Abstract:Motivation: Peptides have attracted the attention in this century due to their remarkable therapeutic properties. Computational tools are being developed to take advantage of existing information, encapsulating knowledge and making it available in a simple way for general public use. However, these are property-specific redundant data systems, and usually do not display the data in a clear way. In some cases, information download is not even possible. This data needs to be available in a simple form for drug design and other biotechnological applications. Results: We developed Peptipedia, a user-friendly database and web application to search, characterise and analyse peptide sequences. Our tool integrates the information from thirty previously reported databases, making it the largest repository of peptides with recorded activities so far. Besides, we implemented a variety of services to increase our tool's usability. The significant differences of our tools with other existing alternatives becomes a substantial contribution to develop biotechnological and bioengineering applications for peptides. Availability: Peptipedia is available for non-commercial use as an open-access software, licensed under the GNU General Public License, version GPL 3.0. The web platform is publicly available at pesb2.cl/peptipedia. Both the source code and sample datasets are available in the GitHub repository https://github.com/CristoferQ/PeptideDatabase. Contact: david.medina@cebib.cl, ana.sanchez@ing.uchile.cl
Combination of digital signal processing and assembled predictive models facilitates the rational design of proteins
Oct 07, 2020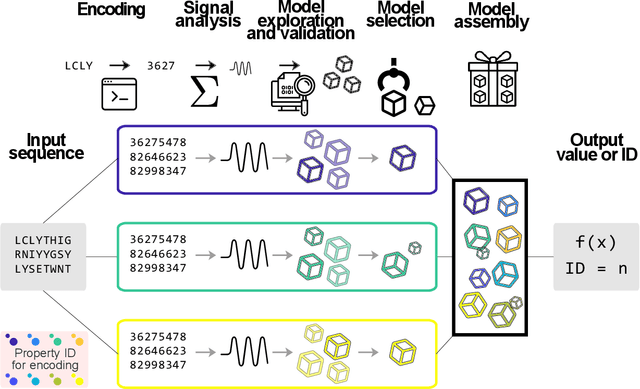
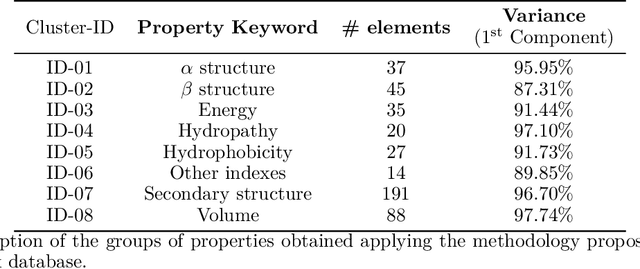
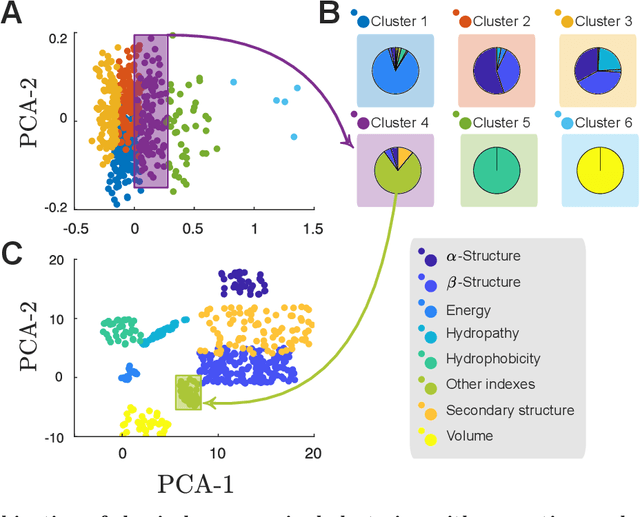
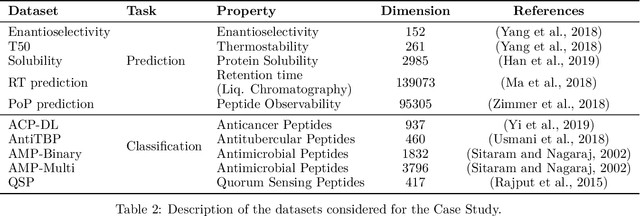
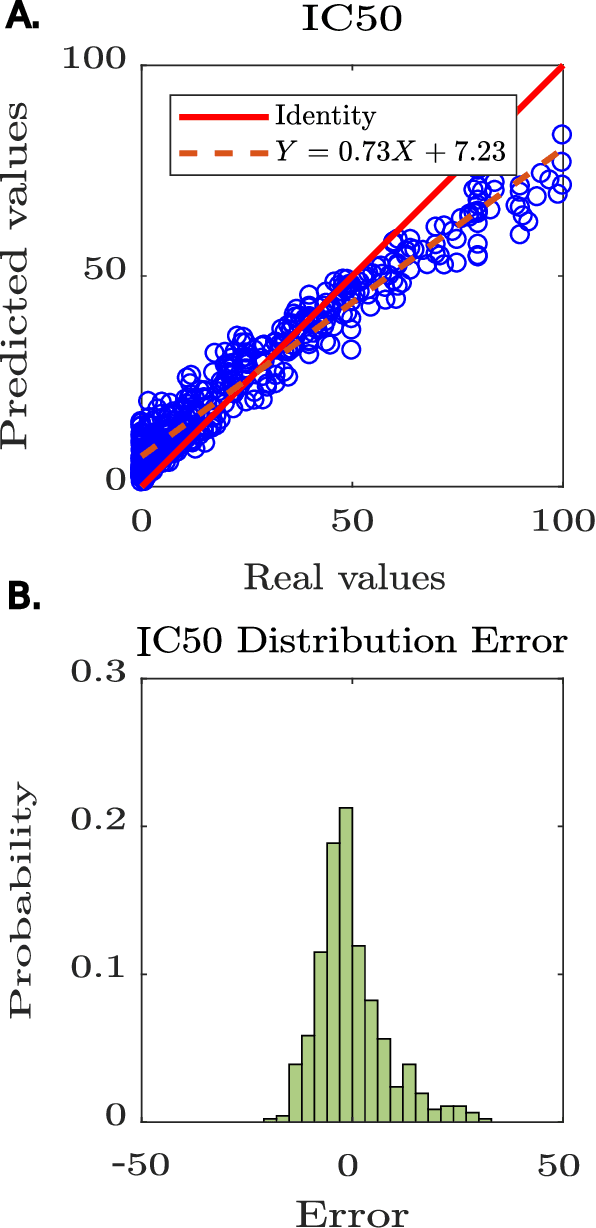
Abstract:Predicting the effect of mutations in proteins is one of the most critical challenges in protein engineering; by knowing the effect a substitution of one (or several) residues in the protein's sequence has on its overall properties, could design a variant with a desirable function. New strategies and methodologies to create predictive models are continually being developed. However, those that claim to be general often do not reach adequate performance, and those that aim to a particular task improve their predictive performance at the cost of the method's generality. Moreover, these approaches typically require a particular decision to encode the amino acidic sequence, without an explicit methodological agreement in such endeavor. To address these issues, in this work, we applied clustering, embedding, and dimensionality reduction techniques to the AAIndex database to select meaningful combinations of physicochemical properties for the encoding stage. We then used the chosen set of properties to obtain several encodings of the same sequence, to subsequently apply the Fast Fourier Transform (FFT) on them. We perform an exploratory stage of Machine-Learning models in the frequency space, using different algorithms and hyperparameters. Finally, we select the best performing predictive models in each set of properties and create an assembled model. We extensively tested the proposed methodology on different datasets and demonstrated that the generated assembled model achieved notably better performance metrics than those models based on a single encoding and, in most cases, better than those previously reported. The proposed method is available as a Python library for non-commercial use under the GNU General Public License (GPLv3) license.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge