David M. Pereira
The chemical space of terpenes: insights from data science and AI
Oct 27, 2021

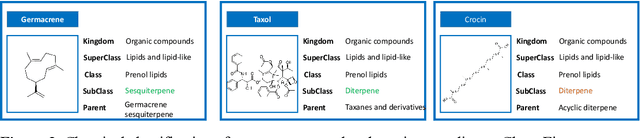

Abstract:Terpenes are a widespread class of natural products with significant chemical and biological diversity and many of these molecules have already made their way into medicines. Given the thousands of molecules already described, the full characterization of this chemical space can be a challenging task when relying in classical approaches. In this work we employ a data science-based approach to identify, compile and characterize the diversity of terpenes currently known in a systematic way. We worked with a natural product database, COCONUT, from which we extracted information for nearly 60000 terpenes. For these molecules, we conducted a subclass-by-subclass analysis in which we highlight several chemical and physical properties relevant to several fields, such as natural products chemistry, medicinal chemistry and drug discovery, among others. We were also interested in assessing the potential of this data for clustering and classification tasks. For clustering, we have applied and compared k-means with agglomerative clustering, both to the original data and following a step of dimensionality reduction. To this end, PCA, FastICA, Kernel PCA, t-SNE and UMAP were used and benchmarked. We also employed a number of methods for the purpose of classifying terpene subclasses using their physico-chemical descriptors. Light gradient boosting machine, k-nearest neighbors, random forests, Gaussian naiive Bayes and Multilayer perceptron, with the best-performing algorithms yielding accuracy, F1 score, precision and other metrics all over 0.9, thus showing the capabilities of these approaches for the classification of terpene subclasses.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge