Daniel Baum
CoDA: Interactive Segmentation and Morphological Analysis of Dendroid Structures Exemplified on Stony Cold-Water Corals
Jun 26, 2024Abstract:Herein, we present CoDA, the Coral Dendroid structure Analyzer, a visual analytics suite that allows for the first time to investigate the ontogenetic morphological development of complex dendroid coral colonies, exemplified on three important framework-forming dendroid cold-water corals: Lophelia pertusa (Linnaeus, 1758), Madrepora oculata (Linnaeus, 1758), and Goniocorella dumosa (Alcock, 1902). Input to CoDA is an initial instance segmentation of the coral polyp cavities (calices), from which it estimates the skeleton tree of the colony and extracts classical morphological measurements and advanced shape features of the individual corallites. CoDA also works as a proofreading and error correction tool by helping to identify wrong parts in the skeleton tree and providing tools to quickly correct these errors. The final skeleton tree enables the derivation of additional information about the calices/corallite instances that otherwise could not be obtained, including their ontogenetic generation and branching patterns - the basis of a fully quantitative statistical analysis of the coral colony morphology. Part of CoDA is CoDAGraph, a feature-rich link-and-brush user interface for visualizing the extracted features and 2D graph layouts of the skeleton tree, enabling the real-time exploration of complex coral colonies and their building blocks, the individual corallites and branches. In the future, we expect CoDA to greatly facilitate the analysis of large stony corals of different species and morphotypes, as well as other dendroid structures, enabling new insights into the influence of genetic and environmental factors on their ontogenetic morphological development.
A Local Iterative Approach for the Extraction of 2D Manifolds from Strongly Curved and Folded Thin-Layer Structures
Aug 14, 2023Abstract:Ridge surfaces represent important features for the analysis of 3-dimensional (3D) datasets in diverse applications and are often derived from varying underlying data including flow fields, geological fault data, and point data, but they can also be present in the original scalar images acquired using a plethora of imaging techniques. Our work is motivated by the analysis of image data acquired using micro-computed tomography (Micro-CT) of ancient, rolled and folded thin-layer structures such as papyrus, parchment, and paper as well as silver and lead sheets. From these documents we know that they are 2-dimensional (2D) in nature. Hence, we are particularly interested in reconstructing 2D manifolds that approximate the document's structure. The image data from which we want to reconstruct the 2D manifolds are often very noisy and represent folded, densely-layered structures with many artifacts, such as ruptures or layer splitting and merging. Previous ridge-surface extraction methods fail to extract the desired 2D manifold for such challenging data. We have therefore developed a novel method to extract 2D manifolds. The proposed method uses a local fast marching scheme in combination with a separation of the region covered by fast marching into two sub-regions. The 2D manifold of interest is then extracted as the surface separating the two sub-regions. The local scheme can be applied for both automatic propagation as well as interactive analysis. We demonstrate the applicability and robustness of our method on both artificial data as well as real-world data including folded silver and papyrus sheets.
A Kendall Shape Space Approach to 3D Shape Estimation from 2D Landmarks
Jul 26, 2022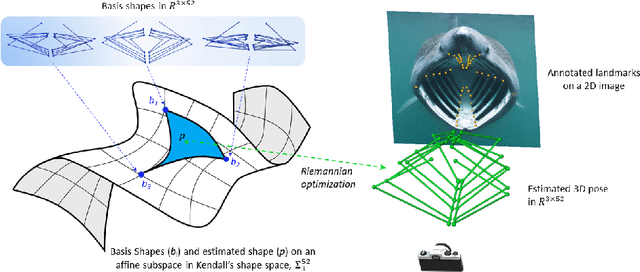
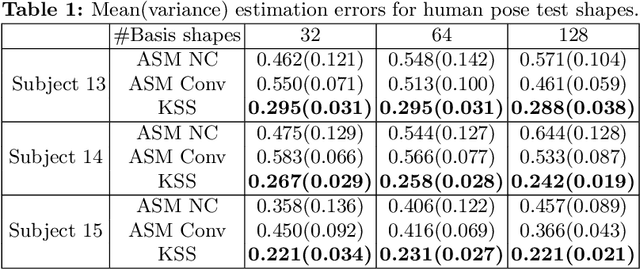
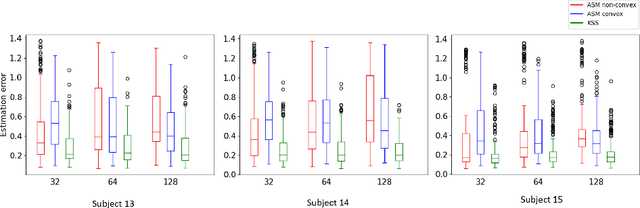
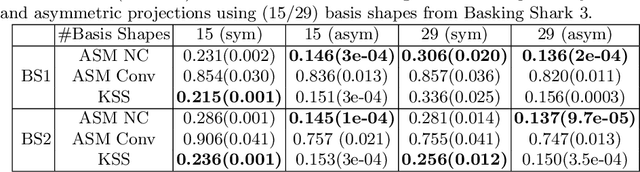
Abstract:3D shapes provide substantially more information than 2D images. However, the acquisition of 3D shapes is sometimes very difficult or even impossible in comparison with acquiring 2D images, making it necessary to derive the 3D shape from 2D images. Although this is, in general, a mathematically ill-posed problem, it might be solved by constraining the problem formulation using prior information. Here, we present a new approach based on Kendall's shape space to reconstruct 3D shapes from single monocular 2D images. The work is motivated by an application to study the feeding behavior of the basking shark, an endangered species whose massive size and mobility render 3D shape data nearly impossible to obtain, hampering understanding of their feeding behaviors and ecology. 2D images of these animals in feeding position, however, are readily available. We compare our approach with state-of-the-art shape-based approaches, both on human stick models and on shark head skeletons. Using a small set of training shapes, we show that the Kendall shape space approach is substantially more robust than previous methods and results in plausible shapes. This is essential for the motivating application in which specimens are rare and therefore only few training shapes are available.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge