Cristiano Russo
NuLite -- Lightweight and Fast Model for Nuclei Instance Segmentation and Classification
Aug 03, 2024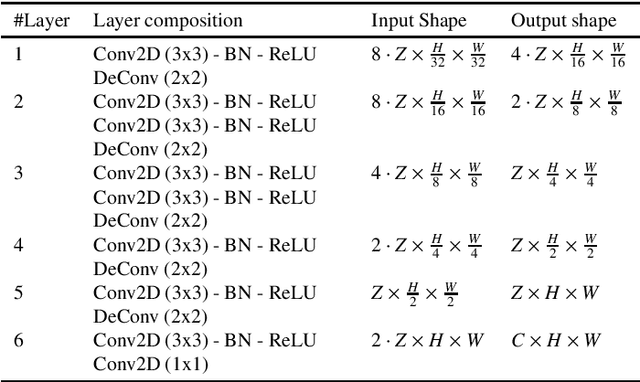


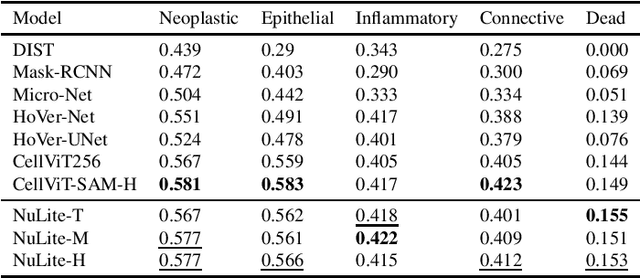
Abstract:In pathology, accurate and efficient analysis of Hematoxylin and Eosin (H\&E) slides is crucial for timely and effective cancer diagnosis. Although many deep learning solutions for nuclei instance segmentation and classification exist in the literature, they often entail high computational costs and resource requirements, thus limiting their practical usage in medical applications. To address this issue, we introduce a novel convolutional neural network, NuLite, a U-Net-like architecture designed explicitly on Fast-ViT, a state-of-the-art (SOTA) lightweight CNN. We obtained three versions of our model, NuLite-S, NuLite-M, and NuLite-H, trained on the PanNuke dataset. The experimental results prove that our models equal CellViT (SOTA) in terms of panoptic quality and detection. However, our lightest model, NuLite-S, is 40 times smaller in terms of parameters and about 8 times smaller in terms of GFlops, while our heaviest model is 17 times smaller in terms of parameters and about 7 times smaller in terms of GFlops. Moreover, our model is up to about 8 times faster than CellViT. Lastly, to prove the effectiveness of our solution, we provide a robust comparison of external datasets, namely CoNseP, MoNuSeg, and GlySAC. Our model is publicly available at https://github.com/CosmoIknosLab/NuLite
HoVer-UNet: Accelerating HoVerNet with UNet-based multi-class nuclei segmentation via knowledge distillation
Dec 04, 2023



Abstract:We present HoVer-UNet, an approach to distill the knowledge of the multi-branch HoVerNet framework for nuclei instance segmentation and classification in histopathology. We propose a compact, streamlined single UNet network with a Mix Vision Transformer backbone, and equip it with a custom loss function to optimally encode the distilled knowledge of HoVerNet, reducing computational requirements without compromising performances. We show that our model achieved results comparable to HoVerNet on the public PanNuke and Consep datasets with a three-fold reduction in inference time. We make the code of our model publicly available at https://github.com/DIAGNijmegen/HoVer-UNet.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge