Christian O'Reilly
SACA: Selective Attention-Based Clustering Algorithm
Aug 23, 2025Abstract:Clustering algorithms are widely used in various applications, with density-based methods such as Density-Based Spatial Clustering of Applications with Noise (DBSCAN) being particularly prominent. These algorithms identify clusters in high-density regions while treating sparser areas as noise. However, reliance on user-defined parameters often poses optimization challenges that require domain expertise. This paper presents a novel density-based clustering method inspired by the concept of selective attention, which minimizes the need for user-defined parameters under standard conditions. Initially, the algorithm operates without requiring user-defined parameters. If parameter adjustment is needed, the method simplifies the process by introducing a single integer parameter that is straightforward to tune. The approach computes a threshold to filter out the most sparsely distributed points and outliers, forms a preliminary cluster structure, and then reintegrates the excluded points to finalize the results. Experimental evaluations on diverse data sets highlight the accessibility and robust performance of the method, providing an effective alternative for density-based clustering tasks.
A Reliable and Efficient Detection Pipeline for Rodent Ultrasonic Vocalizations
Mar 24, 2025Abstract:Analyzing ultrasonic vocalizations (USVs) is crucial for understanding rodents' affective states and social behaviors, but the manual analysis is time-consuming and prone to errors. Automated USV detection systems have been developed to address these challenges. Yet, these systems often rely on machine learning and fail to generalize effectively to new datasets. To tackle these shortcomings, we introduce ContourUSV, an efficient automated system for detecting USVs from audio recordings. Our pipeline includes spectrogram generation, cleaning, pre-processing, contour detection, post-processing, and evaluation against manual annotations. To ensure robustness and reliability, we compared ContourUSV with three state-of-the-art systems using an existing open-access USV dataset (USVSEG) and a second dataset we are releasing publicly along with this paper. On average, across the two datasets, ContourUSV outperformed the other three systems with a 1.51x improvement in precision, 1.17x in recall, 1.80x in F1 score, and 1.49x in specificity while achieving an average speedup of 117.07x.
Benchmarking Deep Jansen-Rit Parameter Inference: An in Silico Study
Jun 07, 2024
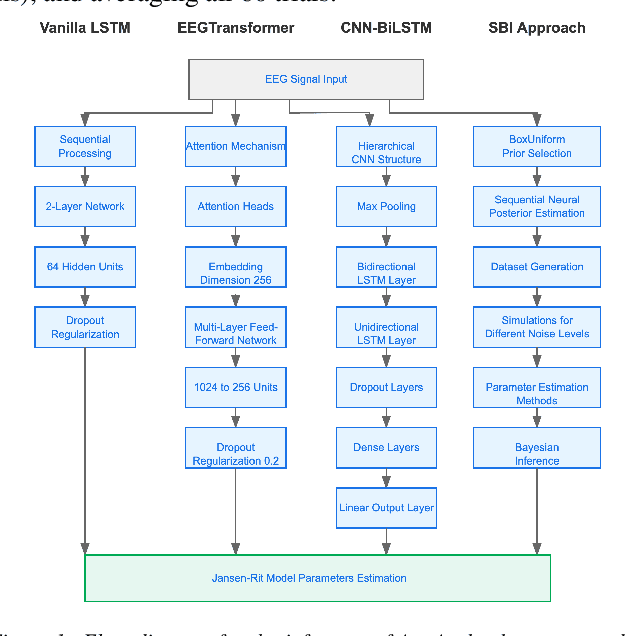


Abstract:The study of effective connectivity (EC) is essential in understanding how the brain integrates and responds to various sensory inputs. Model-driven estimation of EC is a powerful approach that requires estimating global and local parameters of a generative model of neural activity. Insights gathered through this process can be used in various applications, such as studying neurodevelopmental disorders. However, accurately determining EC through generative models remains a significant challenge due to the complexity of brain dynamics and the inherent noise in neural recordings, e.g., in electroencephalography (EEG). Current model-driven methods to study EC are computationally complex and cannot scale to all brain regions as required by whole-brain analyses. To facilitate EC assessment, an inference algorithm must exhibit reliable prediction of parameters in the presence of noise. Further, the relationship between the model parameters and the neural recordings must be learnable. To progress toward these objectives, we benchmarked the performance of a Bi-LSTM model for parameter inference from the Jansen-Rit neural mass model (JR-NMM) simulated EEG under various noise conditions. Additionally, our study explores how the JR-NMM reacts to changes in key biological parameters (i.e., sensitivity analysis) like synaptic gains and time constants, a crucial step in understanding the connection between neural mechanisms and observed brain activity. Our results indicate that we can predict the local JR-NMM parameters from EEG, supporting the feasibility of our deep-learning-based inference approach. In future work, we plan to extend this framework to estimate local and global parameters from real EEG in clinically relevant applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge