Chase Yakaboski
AI for Open Science: A Multi-Agent Perspective for Ethically Translating Data to Knowledge
Oct 31, 2023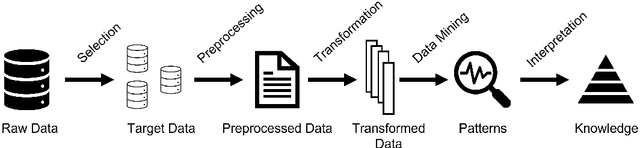
Abstract:AI for Science (AI4Science), particularly in the form of self-driving labs, has the potential to sideline human involvement and hinder scientific discovery within the broader community. While prior research has focused on ensuring the responsible deployment of AI applications, enhancing security, and ensuring interpretability, we also propose that promoting openness in AI4Science discoveries should be carefully considered. In this paper, we introduce the concept of AI for Open Science (AI4OS) as a multi-agent extension of AI4Science with the core principle of maximizing open knowledge translation throughout the scientific enterprise rather than a single organizational unit. We use the established principles of Knowledge Discovery and Data Mining (KDD) to formalize a language around AI4OS. We then discuss three principle stages of knowledge translation embedded in AI4Science systems and detail specific points where openness can be applied to yield an AI4OS alternative. Lastly, we formulate a theoretical metric to assess AI4OS with a supporting ethical argument highlighting its importance. Our goal is that by drawing attention to AI4OS we can ensure the natural consequence of AI4Science (e.g., self-driving labs) is a benefit not only for its developers but for society as a whole.
Learning the Finer Things: Bayesian Structure Learning at the Instantiation Level
Mar 08, 2023Abstract:Successful machine learning methods require a trade-off between memorization and generalization. Too much memorization and the model cannot generalize to unobserved examples. Too much over-generalization and we risk under-fitting the data. While we commonly measure their performance through cross validation and accuracy metrics, how should these algorithms cope in domains that are extremely under-determined where accuracy is always unsatisfactory? We present a novel probabilistic graphical model structure learning approach that can learn, generalize and explain in these elusive domains by operating at the random variable instantiation level. Using Minimum Description Length (MDL) analysis, we propose a new decomposition of the learning problem over all training exemplars, fusing together minimal entropy inferences to construct a final knowledge base. By leveraging Bayesian Knowledge Bases (BKBs), a framework that operates at the instantiation level and inherently subsumes Bayesian Networks (BNs), we develop both a theoretical MDL score and associated structure learning algorithm that demonstrates significant improvements over learned BNs on 40 benchmark datasets. Further, our algorithm incorporates recent off-the-shelf DAG learning techniques enabling tractable results even on large problems. We then demonstrate the utility of our approach in a significantly under-determined domain by learning gene regulatory networks on breast cancer gene mutational data available from The Cancer Genome Atlas (TCGA).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge