Armin Abdehkakha
Effect of structure-based training on 3D localization precision and quality
Sep 29, 2023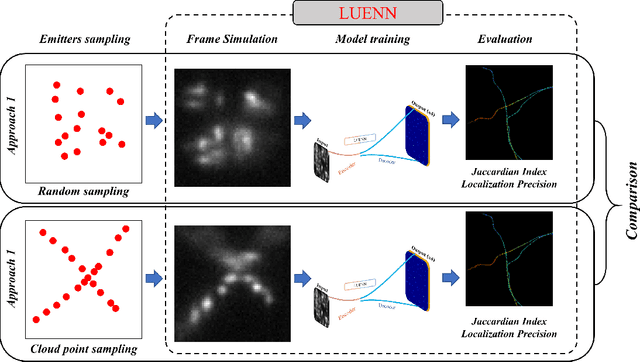
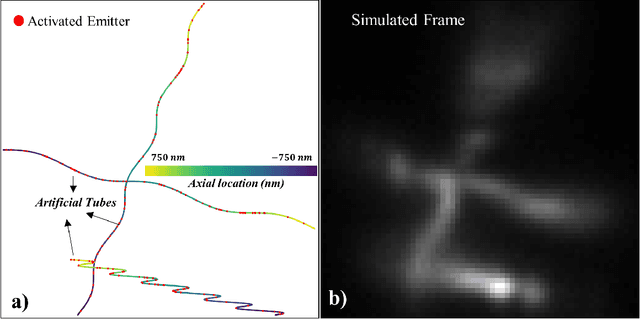
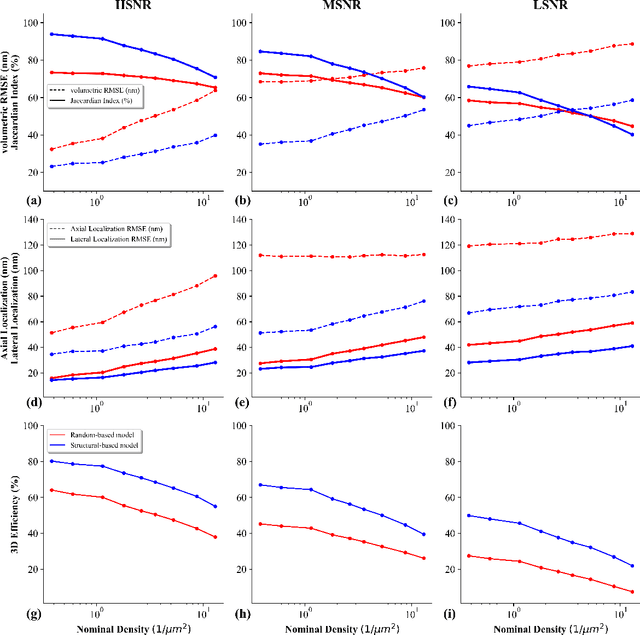
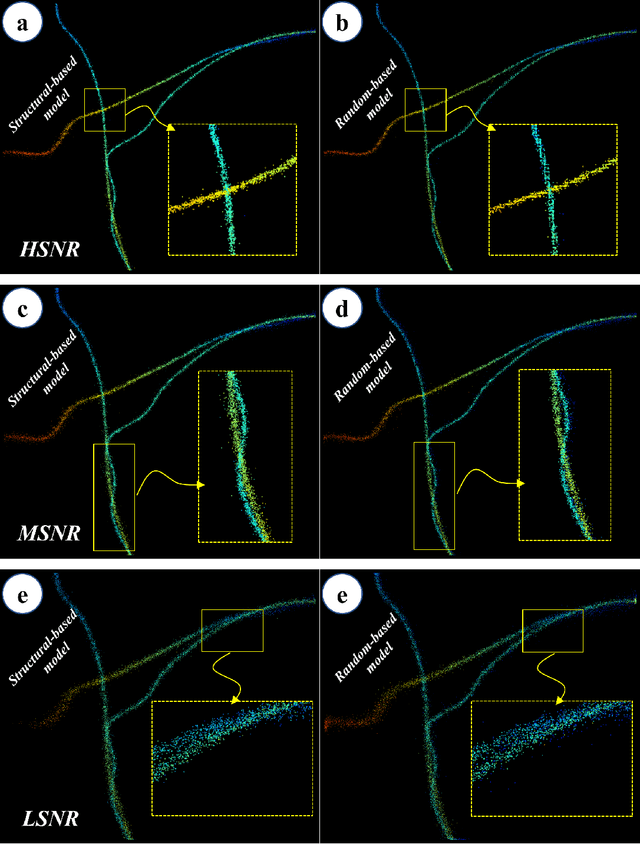
Abstract:This study introduces a structural-based training approach for CNN-based algorithms in single-molecule localization microscopy (SMLM) and 3D object reconstruction. We compare this approach with the traditional random-based training method, utilizing the LUENN package as our AI pipeline. The quantitative evaluation demonstrates significant improvements in detection rate and localization precision with the structural-based training approach, particularly in varying signal-to-noise ratios (SNRs). Moreover, the method effectively removes checkerboard artifacts, ensuring more accurate 3D reconstructions. Our findings highlight the potential of the structural-based training approach to advance super-resolution microscopy and deepen our understanding of complex biological systems at the nanoscale.
Localization of Ultra-dense Emitters with Neural Networks
May 07, 2023Abstract:Single-Molecule Localization Microscopy (SMLM) has expanded our ability to visualize subcellular structures but is limited in its temporal resolution. Increasing emitter density will improve temporal resolution, but current analysis algorithms struggle as emitter images significantly overlap. Here we present a deep convolutional neural network called LUENN which utilizes a unique architecture that rejects the isolated emitter assumption; it can smoothly accommodate emitters that range from completely isolated to co-located. This architecture, alongside an accurate estimator of location uncertainty, extends the range of usable emitter densities by a factor of 6 to over 31 emitters per micrometer-squared with reduced penalty to localization precision and improved temporal resolution. Apart from providing uncertainty estimation, the algorithm improves usability in laboratories by reducing imaging times and easing requirements for successful experiments.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge