Antoine Spahr
ScoreNet: Learning Non-Uniform Attention and Augmentation for Transformer-Based Histopathological Image Classification
Mar 14, 2022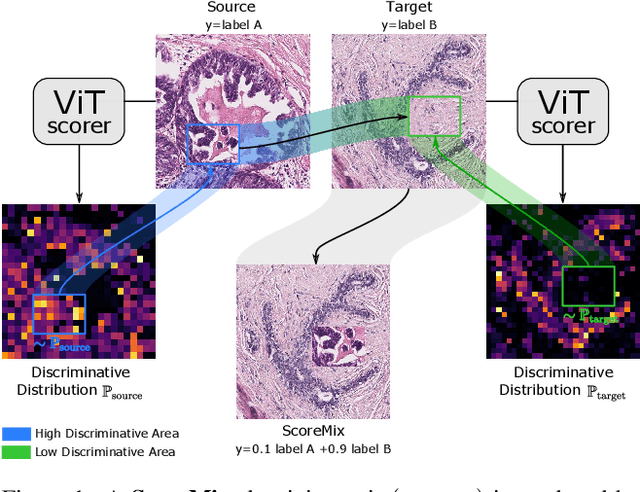

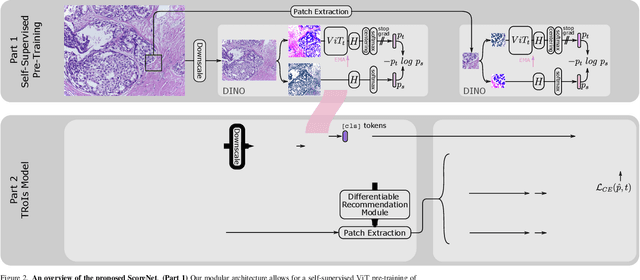

Abstract:Progress in digital pathology is hindered by high-resolution images and the prohibitive cost of exhaustive localized annotations. The commonly used paradigm to categorize pathology images is patch-based processing, which often incorporates multiple instance learning (MIL) to aggregate local patch-level representations yielding image-level prediction. Nonetheless, diagnostically relevant regions may only take a small fraction of the whole tissue, and current MIL-based approaches often process images uniformly, discarding the inter-patches interactions. To alleviate these issues, we propose ScoreNet, a new efficient transformer that exploits a differentiable recommendation stage to extract discriminative image regions and dedicate computational resources accordingly. The proposed transformer leverages the local and global attention of a few dynamically recommended high-resolution regions at an efficient computational cost. We further introduce a novel Mixup-based data-augmentation, namely ScoreMix, by leveraging the image's semantic distribution to guide the data mixing and produce coherent sample-label pairs. ScoreMix is embarrassingly simple and mitigates the pitfalls of previous augmentations, which assume a uniform semantic distribution and risk mislabeling the samples. Thorough experiments and ablation studies on three breast cancer histology datasets of Haematoxylin & Eosin (H&E) have validated the superiority of our approach over prior arts, including transformer-based models on tumour regions-of-interest (TRoIs) classification. ScoreNet equipped with proposed ScoreMix augmentation demonstrates better generalization capabilities and achieves new state-of-the-art (SOTA) results with only 50% of the data compared to other Mixup augmentation variants. Finally, ScoreNet yields high efficacy and outperforms SOTA efficient transformers, namely TransPath and SwinTransformer.
Self-Taught Semi-Supervised Anomaly Detection on Upper Limb X-rays
Feb 22, 2021



Abstract:Detecting anomalies in musculoskeletal radiographs is of paramount importance for large-scale screening in the radiology workflow. Supervised deep networks take for granted a large number of annotations by radiologists, which is often prohibitively very time-consuming to acquire. Moreover, supervised systems are tailored to closed set scenarios, e.g., trained models suffer from overfitting to previously seen rare anomalies at training. Instead, our approach's rationale is to use task agnostic pretext tasks to leverage unlabeled data based on a cross-sample similarity measure. Besides, we formulate a complex distribution of data from normal class within our framework to avoid a potential bias on the side of anomalies. Through extensive experiments, we show that our method outperforms baselines across unsupervised and self-supervised anomaly detection settings on a real-world medical dataset, the MURA dataset. We also provide rich ablation studies to analyze each training stage's effect and loss terms on the final performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge