Andreas Schäfer
FACT: Federated Adversarial Cross Training
Jun 01, 2023



Abstract:Federated Learning (FL) facilitates distributed model development to aggregate multiple confidential data sources. The information transfer among clients can be compromised by distributional differences, i.e., by non-i.i.d. data. A particularly challenging scenario is the federated model adaptation to a target client without access to annotated data. We propose Federated Adversarial Cross Training (FACT), which uses the implicit domain differences between source clients to identify domain shifts in the target domain. In each round of FL, FACT cross initializes a pair of source clients to generate domain specialized representations which are then used as a direct adversary to learn a domain invariant data representation. We empirically show that FACT outperforms state-of-the-art federated, non-federated and source-free domain adaptation models on three popular multi-source-single-target benchmarks, and state-of-the-art Unsupervised Domain Adaptation (UDA) models on single-source-single-target experiments. We further study FACT's behavior with respect to communication restrictions and the number of participating clients.
BITES: Balanced Individual Treatment Effect for Survival data
Jan 05, 2022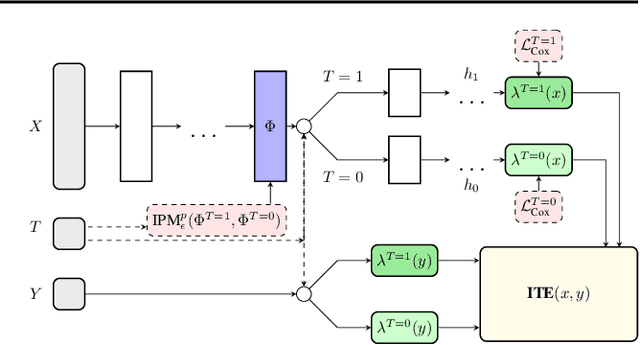
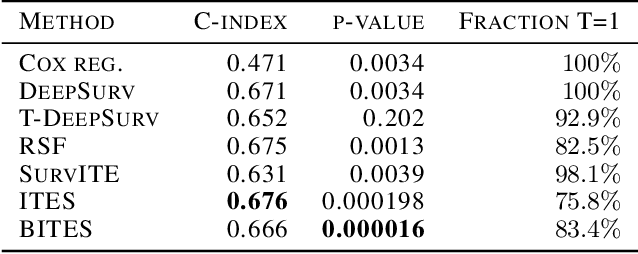
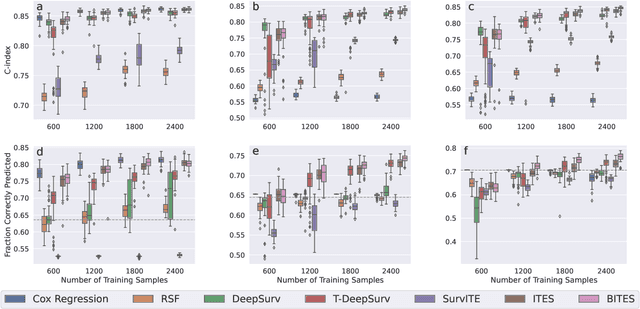
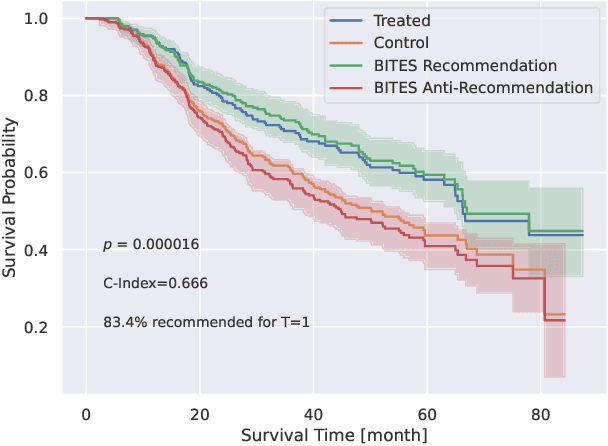
Abstract:Estimating the effects of interventions on patient outcome is one of the key aspects of personalized medicine. Their inference is often challenged by the fact that the training data comprises only the outcome for the administered treatment, and not for alternative treatments (the so-called counterfactual outcomes). Several methods were suggested for this scenario based on observational data, i.e.~data where the intervention was not applied randomly, for both continuous and binary outcome variables. However, patient outcome is often recorded in terms of time-to-event data, comprising right-censored event times if an event does not occur within the observation period. Albeit their enormous importance, time-to-event data is rarely used for treatment optimization. We suggest an approach named BITES (Balanced Individual Treatment Effect for Survival data), which combines a treatment-specific semi-parametric Cox loss with a treatment-balanced deep neural network; i.e.~we regularize differences between treated and non-treated patients using Integral Probability Metrics (IPM). We show in simulation studies that this approach outperforms the state of the art. Further, we demonstrate in an application to a cohort of breast cancer patients that hormone treatment can be optimized based on six routine parameters. We successfully validated this finding in an independent cohort. BITES is provided as an easy-to-use python implementation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge