Ali Mahloojifar
A Deep Bayesian Convolutional Spiking Neural Network-based CAD system with Uncertainty Quantification for Medical Images Classification
Apr 23, 2025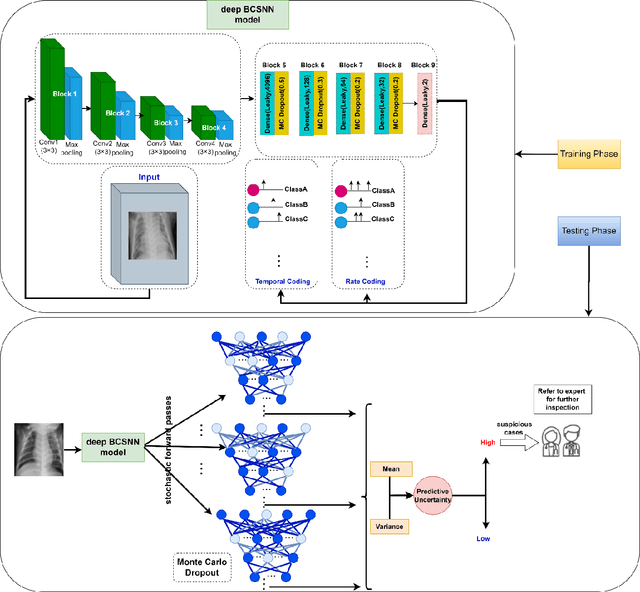



Abstract:The Computer_Aided Diagnosis (CAD) systems facilitate accurate diagnosis of diseases. The development of CADs by leveraging third generation neural network, namely, Spiking Neural Network (SNN), is essential to utilize of the benefits of SNNs, such as their event_driven processing, parallelism, low power consumption, and the ability to process sparse temporal_spatial information. However, Deep SNN as a deep learning model faces challenges with unreliability. To deal with unreliability challenges due to inability to quantify the uncertainty of the predictions, we proposed a deep Bayesian Convolutional Spiking Neural Network based_CADs with uncertainty_aware module. In this study, the Monte Carlo Dropout method as Bayesian approximation is used as an uncertainty quantification method. This method was applied to several medical image classification tasks. Our experimental results demonstrate that our proposed model is accurate and reliable and will be a proper alternative to conventional deep learning for medical image classification.
BI-RADS prediction of mammographic masses using uncertainty information extracted from a Bayesian Deep Learning model
Mar 18, 2025Abstract:The BI_RADS score is a probabilistic reporting tool used by radiologists to express the level of uncertainty in predicting breast cancer based on some morphological features in mammography images. There is a significant variability in describing masses which sometimes leads to BI_RADS misclassification. Using a BI_RADS prediction system is required to support the final radiologist decisions. In this study, the uncertainty information extracted by a Bayesian deep learning model is utilized to predict the BI_RADS score. The investigation results based on the pathology information demonstrate that the f1-scores of the predictions of the radiologist are 42.86%, 48.33% and 48.28%, meanwhile, the f1-scores of the model performance are 73.33%, 59.60% and 59.26% in the BI_RADS 2, 3 and 5 dataset samples, respectively. Also, the model can distinguish malignant from benign samples in the BI_RADS 0 category of the used dataset with an accuracy of 75.86% and correctly identify all malignant samples as BI_RADS 5. The Grad-CAM visualization shows the model pays attention to the morphological features of the lesions. Therefore, this study shows the uncertainty-aware Bayesian Deep Learning model can report his uncertainty about the malignancy of a lesion based on morphological features, like a radiologist.
Reliable Breast Cancer Molecular Subtype Prediction based on uncertainty-aware Bayesian Deep Learning by Mammography
Dec 16, 2024Abstract:Breast cancer is a heterogeneous disease with different molecular subtypes, clinical behavior, treatment responses as well as survival outcomes. The development of a reliable, accurate, available and inexpensive method to predict the molecular subtypes using medical images plays an important role in the diagnosis and prognosis of breast cancer. Recently, deep learning methods have shown good performance in the breast cancer classification tasks using various medical images. Despite all that success, classical deep learning cannot deliver the predictive uncertainty. The uncertainty represents the validity of the predictions.Therefore, the high predicted uncertainty might cause a negative effect in the accurate diagnosis of breast cancer molecular subtypes. To overcome this, uncertainty quantification methods are used to determine the predictive uncertainty. Accordingly, in this study, we proposed an uncertainty-aware Bayesian deep learning model using the full mammogram images. In addition, to increase the performance of the multi-class molecular subtype classification task, we proposed a novel hierarchical classification strategy, named the two-stage classification strategy. The separate AUC of the proposed model for each subtype was 0.71, 0.75 and 0.86 for HER2-enriched, luminal and triple-negative classes, respectively. The proposed model not only has a comparable performance to other studies in the field of breast cancer molecular subtypes prediction, even using full mammography images, but it is also more reliable, due to quantify the predictive uncertainty.
Modifying the U-Net's Encoder-Decoder Architecture for Segmentation of Tumors in Breast Ultrasound Images
Sep 01, 2024Abstract:Segmentation is one of the most significant steps in image processing. Segmenting an image is a technique that makes it possible to separate a digital image into various areas based on the different characteristics of pixels in the image. In particular, segmentation of breast ultrasound images is widely used for cancer identification. As a result of image segmentation, it is possible to make early diagnoses of diseases via medical images in a very effective way. Due to various ultrasound artifacts and noises, including speckle noise, low signal-to-noise ratio, and intensity heterogeneity, the process of accurately segmenting medical images, such as ultrasound images, is still a challenging task. In this paper, we present a new method to improve the accuracy and effectiveness of breast ultrasound image segmentation. More precisely, we propose a Neural Network (NN) based on U-Net and an encoder-decoder architecture. By taking U-Net as the basis, both encoder and decoder parts are developed by combining U-Net with other Deep Neural Networks (Res-Net and MultiResUNet) and introducing a new approach and block (Co-Block), which preserves as much as possible the low-level and the high-level features. The designed network is evaluated using the Breast Ultrasound Images (BUSI) Dataset. It consists of 780 images and the images are categorized into three classes, which are normal, benign, and malignant. According to our extensive evaluations of a public breast ultrasound dataset, the designed network segments the breast lesions more accurately than other state-of-the-art deep learning methods. With only 8.88M parameters, our network (CResU-Net) obtained 76.88%, 71.5%, 90.3%, and 97.4% in terms of Dice similarity coefficients (DSC), Intersection over Union (IoU), Area under curve (AUC), and global accuracy (ACC), respectively, on BUSI dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge