SGC: A semi-supervised pipeline for gene clustering using self-training approach in gene co-expression networks
Paper and Code
Sep 21, 2022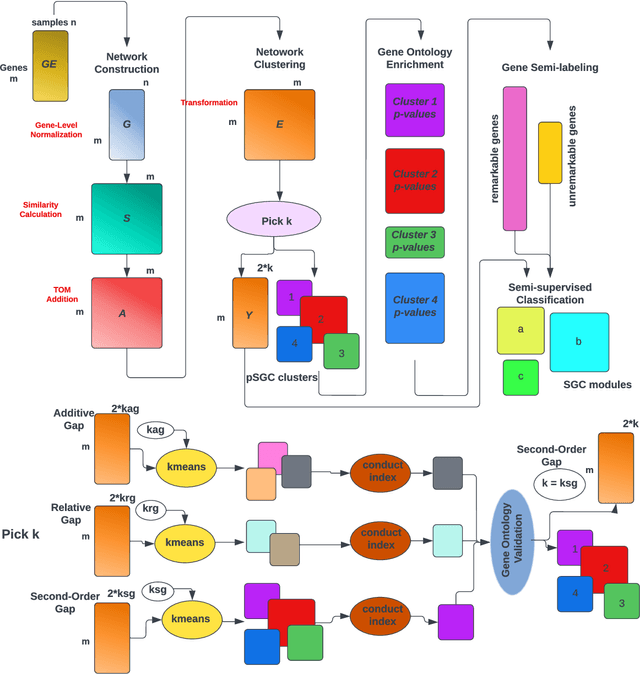
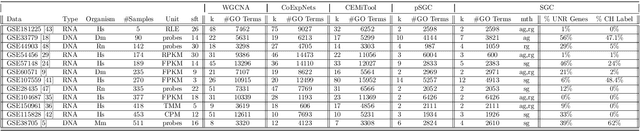
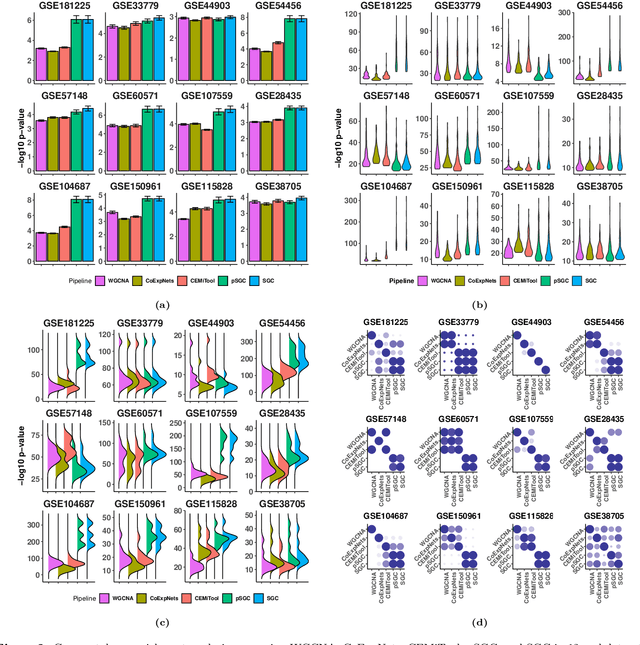
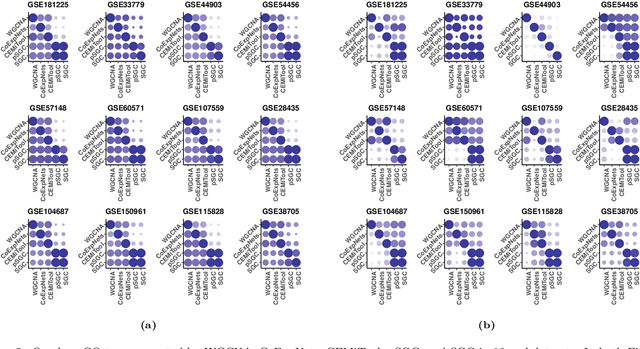
A widely used approach for extracting information from gene expression data employ the construction of a gene co-expression network and the subsequent application of algorithms that discover network structure. In particular, a common goal is the computational discovery of gene clusters, commonly called modules. When applied on a novel gene expression dataset, the quality of the computed modules can be evaluated automatically, using Gene Ontology enrichment, a method that measures the frequencies of Gene Ontology terms in the computed modules and evaluates their statistical likelihood. In this work we propose SGC a novel pipeline for gene clustering based on relatively recent seminal work in the mathematics of spectral network theory. SGC consists of multiple novel steps that enable the computation of highly enriched modules in an unsupervised manner. But unlike all existing frameworks, it further incorporates a novel step that leverages Gene Ontology information in a semi-supervised clustering method that further improves the quality of the computed modules. Comparing with already well-known existing frameworks, we show that SGC results in higher enrichment in real data. In particular, in 12 real gene expression datasets, SGC outperforms in all except one.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge