Optirank: classification for RNA-Seq data with optimal ranking reference genes
Paper and Code
Jan 11, 2023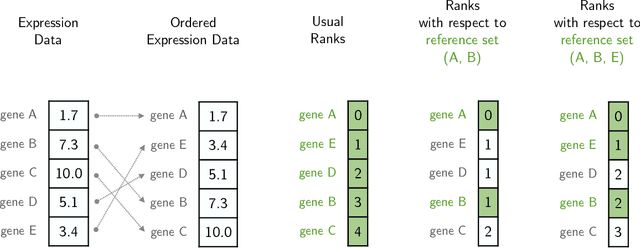

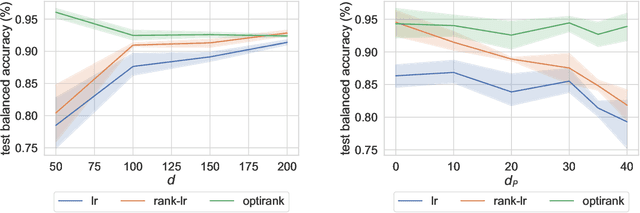
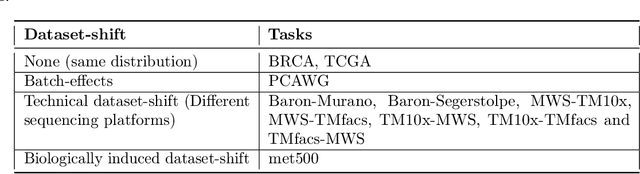
Classification algorithms using RNA-Sequencing (RNA-Seq) data as input are used in a variety of biological applications. By nature, RNA-Seq data is subject to uncontrolled fluctuations both within and especially across datasets, which presents a major difficulty for a trained classifier to generalize to an external dataset. Replacing raw gene counts with the rank of gene counts inside an observation has proven effective to mitigate this problem. However, the rank of a feature is by definition relative to all other features, including highly variable features that introduce noise in the ranking. To address this problem and obtain more robust ranks, we propose a logistic regression model, optirank, which learns simultaneously the parameters of the model and the genes to use as a reference set in the ranking. We show the effectiveness of this method on simulated data. We also consider real classification tasks, which present different kinds of distribution shifts between train and test data. Those tasks concern a variety of applications, such as cancer of unknown primary classification, identification of specific gene signatures, and determination of cell type in single-cell RNA-Seq datasets. On those real tasks, optirank performs at least as well as the vanilla logistic regression on classical ranks, while producing sparser solutions. In addition, to increase the robustness against dataset shifts, we propose a multi-source learning scheme and demonstrate its effectiveness when used in combination with rank-based classifiers.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge