Locally Linear Embedding and fMRI feature selection in psychiatric classification
Paper and Code
Sep 06, 2019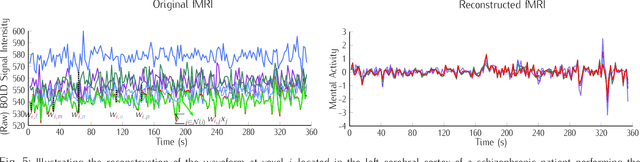
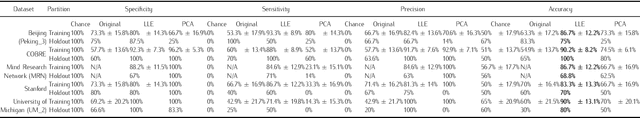
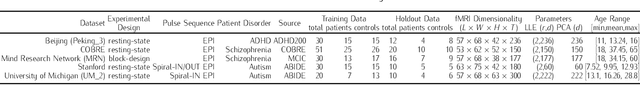
Background: Functional magnetic resonance imaging (fMRI) provides non-invasive measures of neuronal activity using an endogenous Blood Oxygenation-Level Dependent (BOLD) contrast. This article introduces a nonlinear dimensionality reduction (Locally Linear Embedding) to extract informative measures of the underlying neuronal activity from BOLD time-series. The method is validated using the Leave-One-Out-Cross-Validation (LOOCV) accuracy of classifying psychiatric diagnoses using resting-state and task-related fMRI. Methods: Locally Linear Embedding of BOLD time-series (into each voxel's respective tensor) was used to optimise feature selection. This uses Gau\ss' Principle of Least Constraint to conserve quantities over both space and time. This conservation was assessed using LOOCV to greedily select time points in an incremental fashion on training data that was categorised in terms of psychiatric diagnoses. Findings: The embedded fMRI gave highly diagnostic performances (> 80%) on eleven publicly-available datasets containing healthy controls and patients with either Schizophrenia, Attention-Deficit Hyperactivity Disorder (ADHD), or Autism Spectrum Disorder (ASD). Furthermore, unlike the original fMRI data before or after using Principal Component Analysis (PCA) for artefact reduction, the embedded fMRI furnished significantly better than chance classification (defined as the majority class proportion) on ten of eleven datasets Interpretation: Locally Linear Embedding appears to be a useful feature extraction procedure that retains important information about patterns of brain activity distinguishing among psychiatric cohorts.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge