Learning local discrete features in explainable-by-design convolutional neural networks
Paper and Code
Oct 31, 2024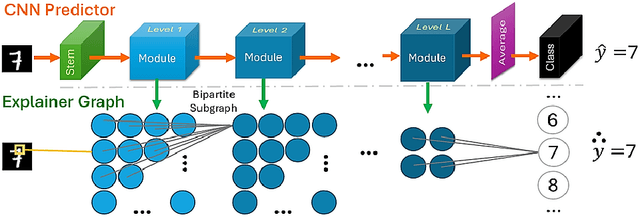

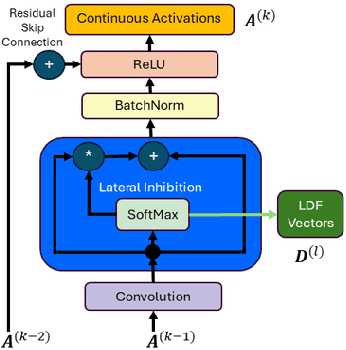

Our proposed framework attempts to break the trade-off between performance and explainability by introducing an explainable-by-design convolutional neural network (CNN) based on the lateral inhibition mechanism. The ExplaiNet model consists of the predictor, that is a high-accuracy CNN with residual or dense skip connections, and the explainer probabilistic graph that expresses the spatial interactions of the network neurons. The value on each graph node is a local discrete feature (LDF) vector, a patch descriptor that represents the indices of antagonistic neurons ordered by the strength of their activations, which are learned with gradient descent. Using LDFs as sequences we can increase the conciseness of explanations by repurposing EXTREME, an EM-based sequence motif discovery method that is typically used in molecular biology. Having a discrete feature motif matrix for each one of intermediate image representations, instead of a continuous activation tensor, allows us to leverage the inherent explainability of Bayesian networks. By collecting observations and directly calculating probabilities, we can explain causal relationships between motifs of adjacent levels and attribute the model's output to global motifs. Moreover, experiments on various tiny image benchmark datasets confirm that our predictor ensures the same level of performance as the baseline architecture for a given count of parameters and/or layers. Our novel method shows promise to exceed this performance while providing an additional stream of explanations. In the solved MNIST classification task, it reaches a comparable to the state-of-the-art performance for single models, using standard training setup and 0.75 million parameters.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge