Implicit Shape Modeling for Anatomical Structure Refinement of Volumetric Medical Images
Paper and Code
Dec 11, 2023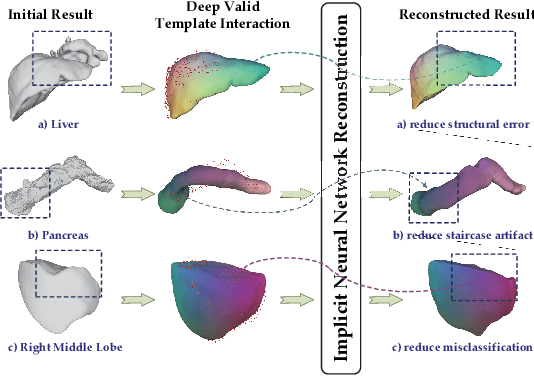
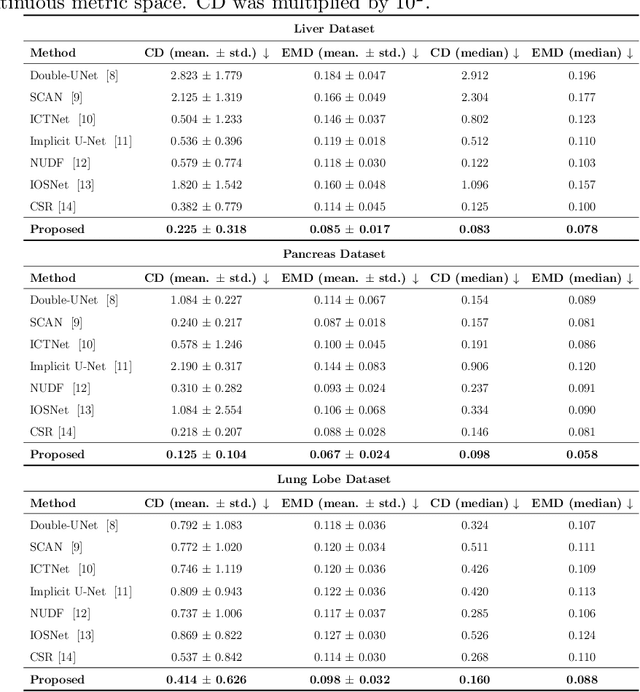
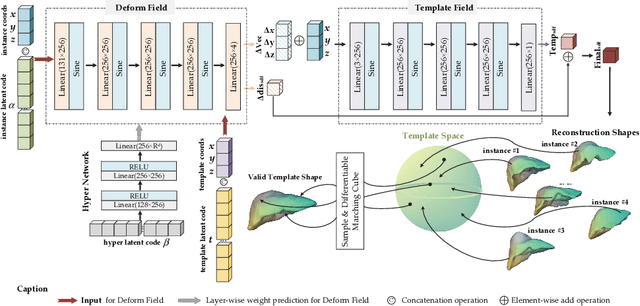
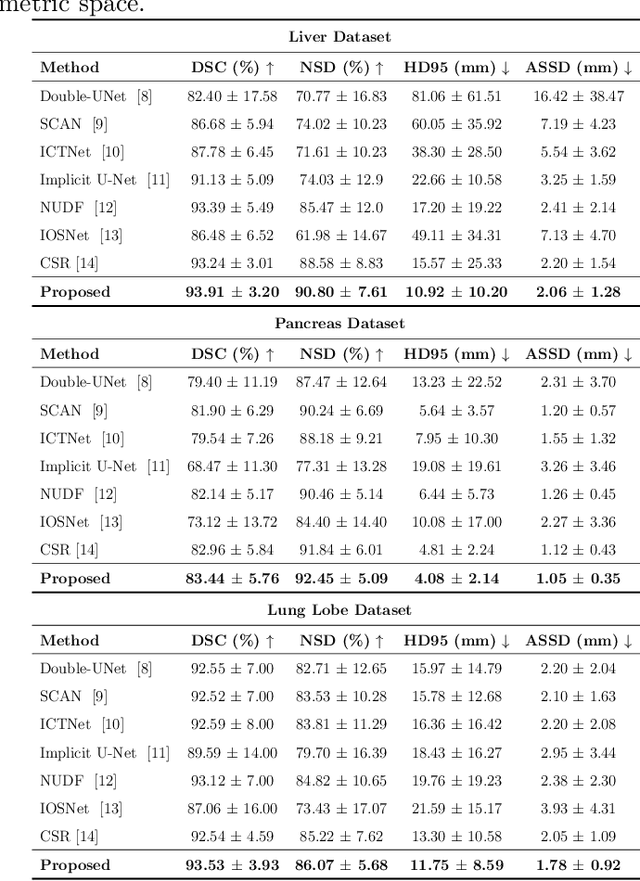
Shape modeling of volumetric medical images is a critical task for quantitative analysis and surgical plans in computer-aided diagnosis. To relieve the burden of expert clinicians, the reconstructed shapes are widely acquired from deep learning models, e.g. Convolutional Neural Networks (CNNs), followed by marching cube algorithm. However, automatically obtaining reconstructed shapes can not always achieve perfect results due to the limited resolution of images and lack of shape prior constraints. In this paper, we design a unified framework for the refinement of medical image segmentation on top of an implicit neural network. Specifically, To learn a sharable shape prior from different instances within the same category in the training phase, the physical information of volumetric medical images are firstly utilized to construct the Physical-Informed Continuous Coordinate Transform (PICCT). PICCT transforms the input data in an aligned manner fed into the implicit shape modeling. To better learn shape representation, we introduce implicit shape constraints on top of the signed distance function (SDF) into the implicit shape modeling of both instances and latent template. For the inference phase, a template interaction module (TIM) is proposed to refine initial results produced by CNNs via deforming deep implicit templates with latent codes. Experimental results on three datasets demonstrated the superiority of our approach in shape refinement. The Chamfer Distance/Earth Mover's Distance achieved by the proposed method are 0.232/0.087 on the Liver dataset, 0.128/0.069 on the Pancreas dataset, and 0.417/0.100 on the Lung Lobe dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge