Implanting Synthetic Lesions for Improving Liver Lesion Segmentation in CT Exams
Paper and Code
Aug 11, 2020
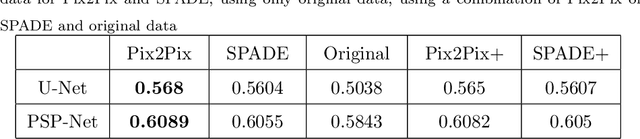
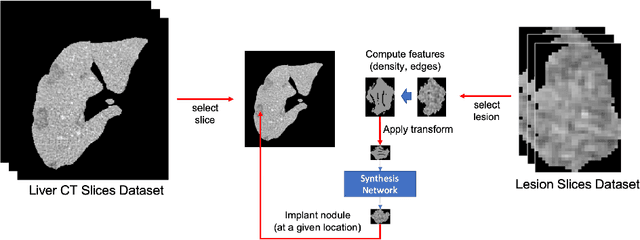
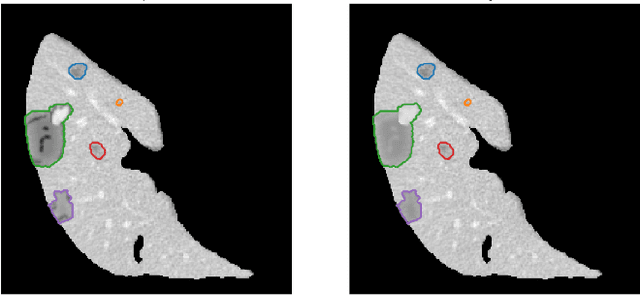
The success of supervised lesion segmentation algorithms using Computed Tomography (CT) exams depends significantly on the quantity and variability of samples available for training. While annotating such data constitutes a challenge itself, the variability of lesions in the dataset also depends on the prevalence of different types of lesions. This phenomenon adds an inherent bias to lesion segmentation algorithms that can be diminished, among different possibilities, using aggressive data augmentation methods. In this paper, we present a method for implanting realistic lesions in CT slices to provide a rich and controllable set of training samples and ultimately improving semantic segmentation network performances for delineating lesions in CT exams. Our results show that implanting synthetic lesions not only improves (up to around 12\%) the segmentation performance considering different architectures but also that this improvement is consistent among different image synthesis networks. We conclude that increasing the variability of lesions synthetically in terms of size, density, shape, and position seems to improve the performance of segmentation models for liver lesion segmentation in CT slices.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge