Hypergraph and protein function prediction with gene expression data
Paper and Code
Dec 03, 2012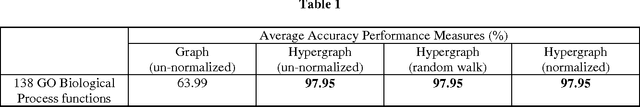
Most network-based protein (or gene) function prediction methods are based on the assumption that the labels of two adjacent proteins in the network are likely to be the same. However, assuming the pairwise relationship between proteins or genes is not complete, the information a group of genes that show very similar patterns of expression and tend to have similar functions (i.e. the functional modules) is missed. The natural way overcoming the information loss of the above assumption is to represent the gene expression data as the hypergraph. Thus, in this paper, the three un-normalized, random walk, and symmetric normalized hypergraph Laplacian based semi-supervised learning methods applied to hypergraph constructed from the gene expression data in order to predict the functions of yeast proteins are introduced. Experiment results show that the average accuracy performance measures of these three hypergraph Laplacian based semi-supervised learning methods are the same. However, their average accuracy performance measures of these three methods are much greater than the average accuracy performance measures of un-normalized graph Laplacian based semi-supervised learning method (i.e. the baseline method of this paper) applied to gene co-expression network created from the gene expression data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge