Gene Transformer: Transformers for the Gene Expression-based Classification of Cancer Subtypes
Paper and Code
Aug 26, 2021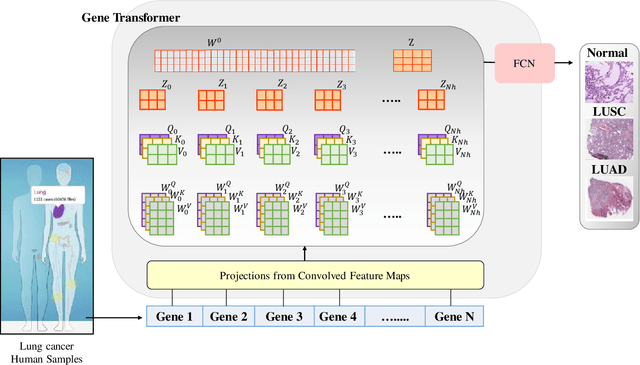

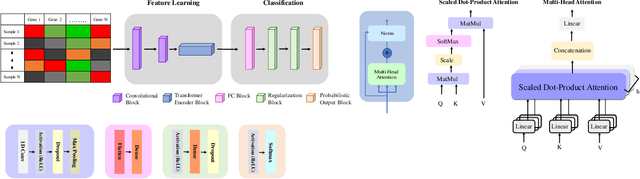

Adenocarcinoma and squamous cell carcinoma constitute approximately 40% and 30% of all lung cancer subtypes, respectively, and display broad heterogeneity in terms of clinical and molecular responses to therapy. Molecular subtyping has enabled precision medicine to overcome these challenges and provide significant biological insights to predict prognosis and improve clinical decision making. Over the past decade, conventional ML algorithms and DL-based CNNs have been espoused for the classification of cancer subtypes from gene expression datasets. However, these methods are potentially biased toward identification of cancer biomarkers. Recently proposed transformer-based architectures that leverage the self-attention mechanism encode high throughput gene expressions and learn representations that are computationally complex and parametrically expensive. However, compared to the datasets for natural language processing applications, gene expression consists of several hundreds of thousands of genes from a limited number of observations, making it difficult to efficiently train transformers for bioinformatics applications. Hence, we propose an end-to-end deep learning approach, Gene Transformer, which addresses the complexity of high-dimensional gene expression with a multi-head self-attention module by identifying relevant biomarkers across multiple cancer subtypes without requiring feature selection as a prerequisite for the current classification algorithms. The proposed architecture achieved an overall improved performance for all evaluation metrics and had fewer misclassification errors than the commonly used traditional classification algorithms. The classification results show that Gene Transformer can be an efficient approach for classifying cancer subtypes, indicating that any improvement in deep learning models in computational biology can also be reflected well in this domain.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge