FunQG: Molecular Representation Learning Via Quotient Graphs
Paper and Code
Jul 18, 2022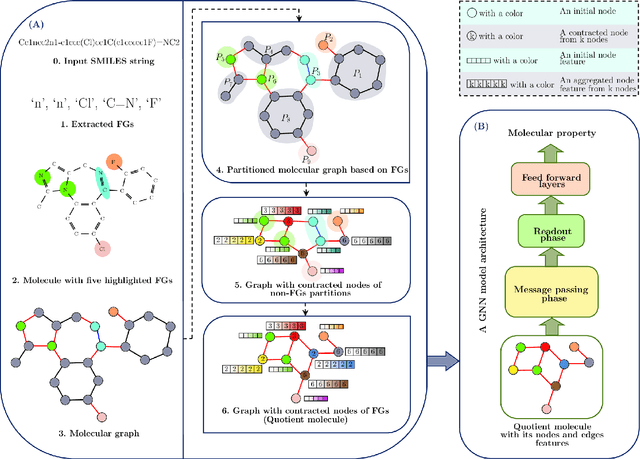
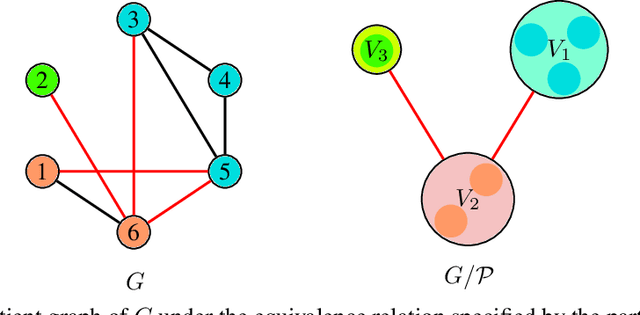
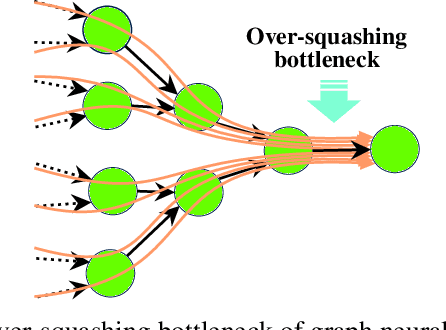
Learning expressive molecular representations is crucial to facilitate the accurate prediction of molecular properties. Despite the significant advancement of graph neural networks (GNNs) in molecular representation learning, they generally face limitations such as neighbors-explosion, under-reaching, over-smoothing, and over-squashing. Also, GNNs usually have high computational complexity because of the large-scale number of parameters. Typically, such limitations emerge or increase when facing relatively large-size graphs or using a deeper GNN model architecture. An idea to overcome these problems is to simplify a molecular graph into a small, rich, and informative one, which is more efficient and less challenging to train GNNs. To this end, we propose a novel molecular graph coarsening framework named FunQG utilizing Functional groups, as influential building blocks of a molecule to determine its properties, based on a graph-theoretic concept called Quotient Graph. By experiments, we show that the resulting informative graphs are much smaller than the molecular graphs and thus are good candidates for training GNNs. We apply the FunQG on popular molecular property prediction benchmarks and then compare the performance of a GNN architecture on the obtained datasets with several state-of-the-art baselines on the original datasets. By experiments, this method significantly outperforms previous baselines on various datasets, besides its dramatic reduction in the number of parameters and low computational complexity. Therefore, the FunQG can be used as a simple, cost-effective, and robust method for solving the molecular representation learning problem.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge