Diagnosis and Analysis of Celiac Disease and Environmental Enteropathy on Biopsy Images using Deep Learning Approaches
Paper and Code
Jun 11, 2020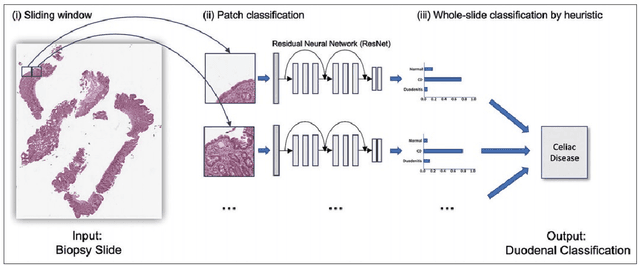
Celiac Disease (CD) and Environmental Enteropathy (EE) are common causes of malnutrition and adversely impact normal childhood development. Both conditions require a tissue biopsy for diagnosis and a major challenge of interpreting clinical biopsy images to differentiate between these gastrointestinal diseases is striking histopathologic overlap between them. In the current study, we propose four diagnosis techniques for these diseases and address their limitations and advantages. First, the diagnosis between CD, EE, and Normal biopsies is considered, but the main challenge with this diagnosis technique is the staining problem. The dataset used in this research is collected from different centers with different staining standards. To solve this problem, we use color balancing in order to train our model with a varying range of colors. Random Multimodel Deep Learning (RMDL) architecture has been used as another approach to mitigate the effects of the staining problem. RMDL combines different architectures and structures of deep learning and the final output of the model is based on the majority vote. CD is a chronic autoimmune disease that affects the small intestine genetically predisposed children and adults. Typically, CD rapidly progress from Marsh I to IIIa. Marsh III is sub-divided into IIIa (partial villus atrophy), Marsh IIIb (subtotal villous atrophy), and Marsh IIIc (total villus atrophy) to explain the spectrum of villus atrophy along with crypt hypertrophy and increased intraepithelial lymphocytes. In the second part of this study, we proposed two ways for diagnosing different stages of CD. Finally, in the third part of this study, these two steps are combined as Hierarchical Medical Image Classification (HMIC) to have a model to diagnose the disease data hierarchically.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge