Deep Multi-attribute Graph Representation Learning on Protein Structures
Paper and Code
Dec 22, 2020
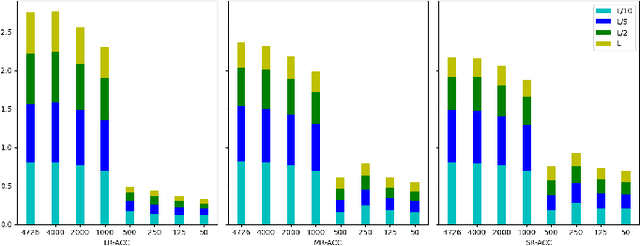


Graphs as a type of data structure have recently attracted significant attention. Representation learning of geometric graphs has achieved great success in many fields including molecular, social, and financial networks. It is natural to present proteins as graphs in which nodes represent the residues and edges represent the pairwise interactions between residues. However, 3D protein structures have rarely been studied as graphs directly. The challenges include: 1) Proteins are complex macromolecules composed of thousands of atoms making them much harder to model than micro-molecules. 2) Capturing the long-range pairwise relations for protein structure modeling remains under-explored. 3) Few studies have focused on learning the different attributes of proteins together. To address the above challenges, we propose a new graph neural network architecture to represent the proteins as 3D graphs and predict both distance geometric graph representation and dihedral geometric graph representation together. This gives a significant advantage because this network opens a new path from the sequence to structure. We conducted extensive experiments on four different datasets and demonstrated the effectiveness of the proposed method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge