Deep Metric Learning with Locality Sensitive Angular Loss for Self-Correcting Source Separation of Neural Spiking Signals
Paper and Code
Oct 13, 2021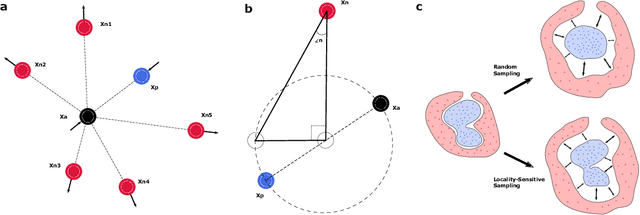
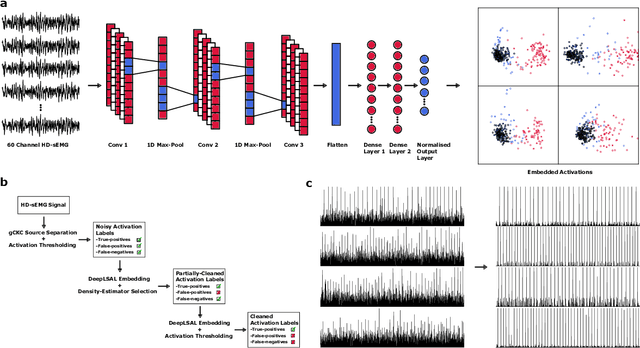
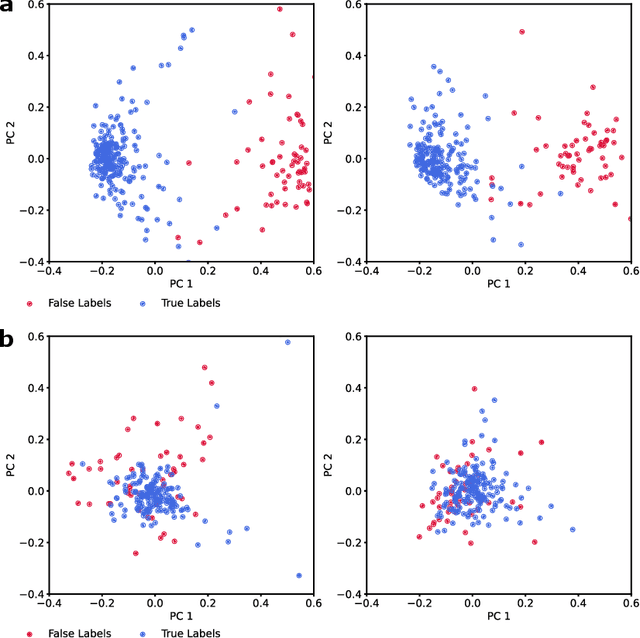
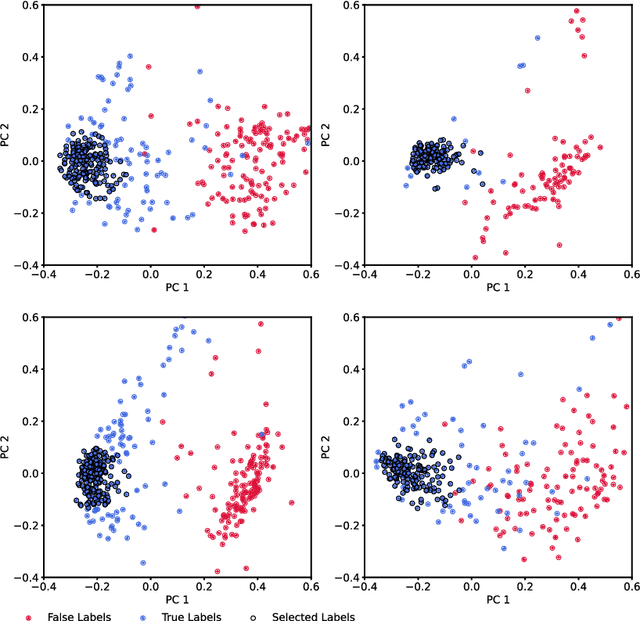
Neurophysiological time series, such as electromyographic signal and intracortical recordings, are typically composed of many individual spiking sources, the recovery of which can give fundamental insights into the biological system of interest or provide neural information for man-machine interfaces. For this reason, source separation algorithms have become an increasingly important tool in neuroscience and neuroengineering. However, in noisy or highly multivariate recordings these decomposition techniques often make a large number of errors, which degrades human-machine interfacing applications and often requires costly post-hoc manual cleaning of the output label set of spike timestamps. To address both the need for automated post-hoc cleaning and robust separation filters we propose a methodology based on deep metric learning, using a novel loss function which maintains intra-class variance, creating a rich embedding space suitable for both label cleaning and the discovery of new activations. We then validate this method with an artificially corrupted label set based on source-separated high-density surface electromyography recordings, recovering the original timestamps even in extreme degrees of feature and class-dependent label noise. This approach enables a neural network to learn to accurately decode neurophysiological time series using any imperfect method of labelling the signal.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge