Comparison of 2D vs. 3D U-Net Organ Segmentation in abdominal 3D CT images
Paper and Code
Jul 08, 2021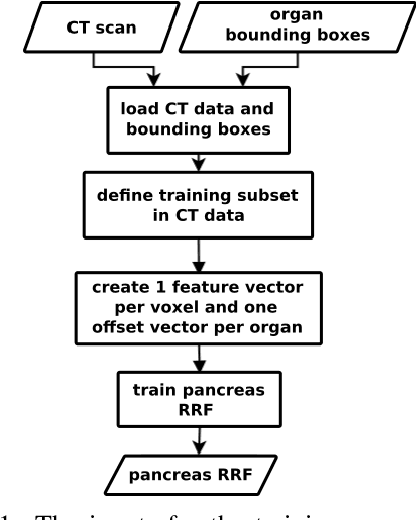
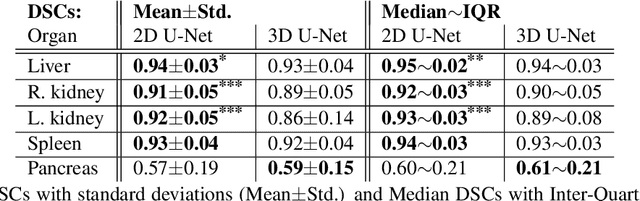
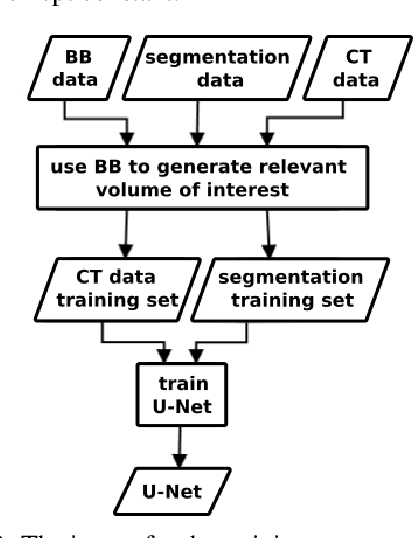
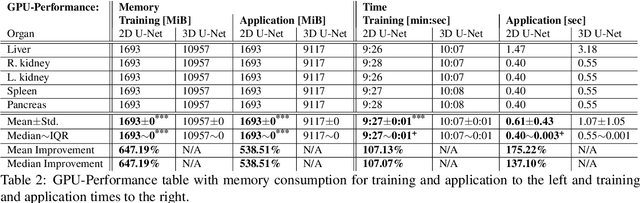
A two-step concept for 3D segmentation on 5 abdominal organs inside volumetric CT images is presented. First each relevant organ's volume of interest is extracted as bounding box. The extracted volume acts as input for a second stage, wherein two compared U-Nets with different architectural dimensions re-construct an organ segmentation as label mask. In this work, we focus on comparing 2D U-Nets vs. 3D U-Net counterparts. Our initial results indicate Dice improvements of about 6\% at maximum. In this study to our surprise, liver and kidneys for instance were tackled significantly better using the faster and GPU-memory saving 2D U-Nets. For other abdominal key organs, there were no significant differences, but we observe highly significant advantages for the 2D U-Net in terms of GPU computational efforts for all organs under study.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge