Augmenting Molecular Images with Vector Representations as a Featurization Technique for Drug Classification
Paper and Code
Aug 09, 2020
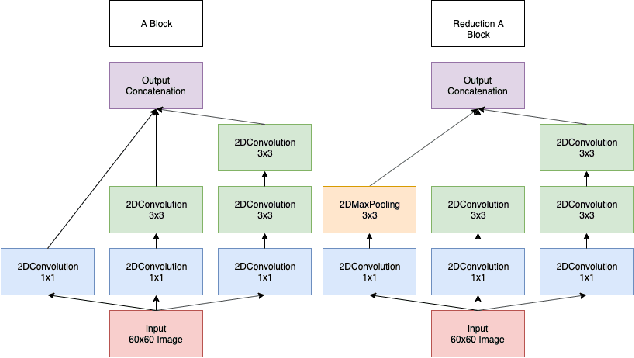
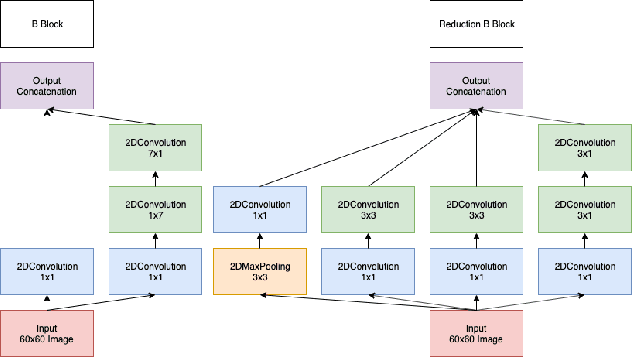
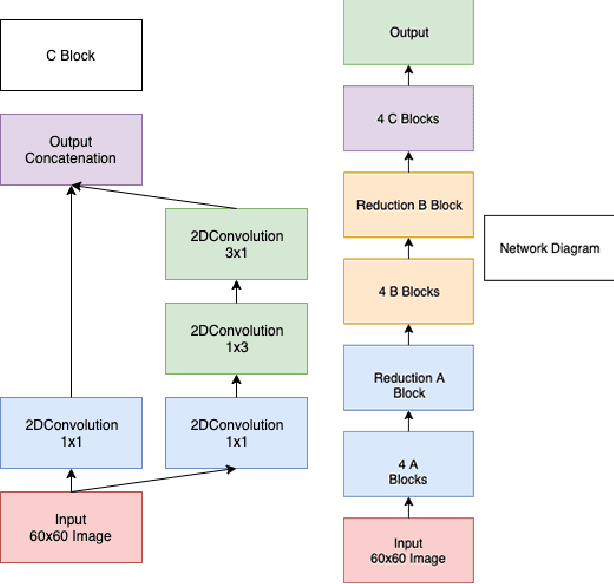
One of the key steps in building deep learning systems for drug classification and generation is the choice of featurization for the molecules. Previous featurization methods have included molecular images, binary strings, graphs, and SMILES strings. This paper proposes the creation of molecular images captioned with binary vectors that encode information not contained in or easily understood from a molecular image alone. Specifically, we use Morgan fingerprints, which encode higher level structural information, and MACCS keys, which encode yes or no questions about a molecules properties and structure. We tested our method on the HIV dataset published by the Pande lab, which consists of 41,127 molecules labeled by if they inhibit the HIV virus. Our final model achieved a state of the art AUC ROC on the HIV dataset, outperforming all other methods. Moreover, the model converged significantly faster than most other methods, requiring dramatically less computational power than unaugmented images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge