A Constraint Programming Approach to Weighted Isomorphic Mapping of Fragment-based Shape Signatures
Paper and Code
Jan 04, 2022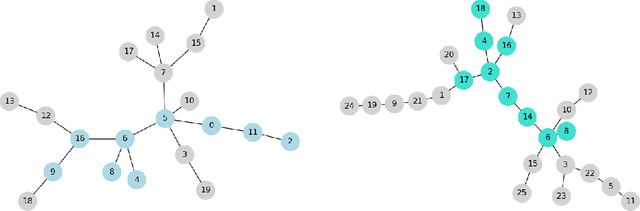
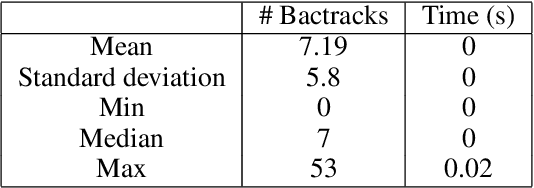
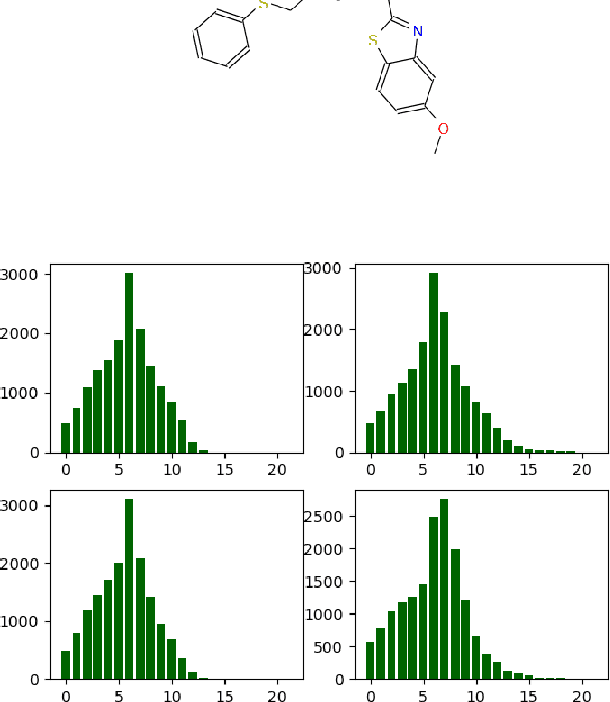
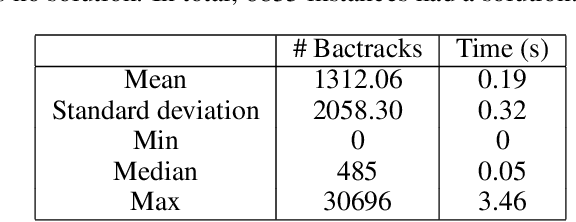
Fragment-based shape signature techniques have proven to be powerful tools for computer-aided drug design. They allow scientists to search for target molecules with some similarity to a known active compound. They do not require reference to the full underlying chemical structure, which is essential to deal with chemical databases containing millions of compounds. However, finding the optimal match of a part of the fragmented compound can be time-consuming. In this paper, we use constraint programming to solve this specific problem. It involves finding a weighted assignment of fragments subject to connectivity constraints. Our experiments demonstrate the practical relevance of our approach and open new perspectives, including generating multiple, diverse solutions. Our approach constitutes an original use of a constraint solver in a real time setting, where propagation allows to avoid an enumeration of weighted paths. The model must remain robust to the addition of constraints making some instances not tractable. This particular context requires the use of unusual criteria for the choice of the model: lightweight, standard propagation algorithms, data structures without prohibitive constant cost. The objective is not to design new, complex algorithms to solve difficult instances.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge